MOF Suppresses Replication Stress and Contributes to Resolution of Stalled Replication Forks
- PMID: 29298824
- PMCID: PMC5829482
- DOI: 10.1128/MCB.00484-17
MOF Suppresses Replication Stress and Contributes to Resolution of Stalled Replication Forks
Abstract
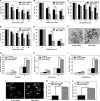
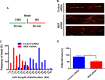
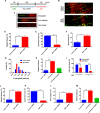
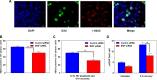
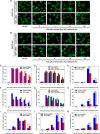
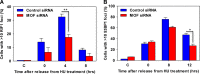
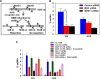
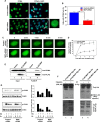
The human MOF (hMOF) protein belongs to the MYST family of histone acetyltransferases and plays a critical role in transcription and the DNA damage response. MOF is essential for cell proliferation; however, its role during replication and replicative stress is unknown. Here we demonstrate that cells depleted of MOF and under replicative stress induced by cisplatin, hydroxyurea, or camptothecin have reduced survival, a higher frequency of S-phase-specific chromosome damage, and increased R-loop formation. MOF depletion decreased replication fork speed and, when combined with replicative stress, also increased stalled replication forks as well as new origin firing. MOF interacted with PCNA, a key coordinator of replication and repair machinery at replication forks, and affected its ubiquitination and recruitment to the DNA damage site. Depletion of MOF, therefore, compromised the DNA damage repair response as evidenced by decreased Mre11, RPA70, Rad51, and PCNA focus formation, reduced DNA end resection, and decreased CHK1 phosphorylation in cells after exposure to hydroxyurea or cisplatin. These results support the argument that MOF plays an important role in suppressing replication stress induced by genotoxic agents at several stages during the DNA damage response.
Keywords: MOF; PCNA; R loop; homologous recombination; replication stress.
Copyright © 2018 American Society for Microbiology.
Figures
Similar articles
-
Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest.Mol Cell Biol. 2013 Mar;33(6):1210-22. doi: 10.1128/MCB.01676-12. Epub 2013 Jan 14. Mol Cell Biol. 2013. PMID: 23319047 Free PMC article.
-
Pol eta is required for DNA replication during nucleotide deprivation by hydroxyurea.Oncogene. 2007 Aug 23;26(39):5713-21. doi: 10.1038/sj.onc.1210385. Epub 2007 Mar 19. Oncogene. 2007. PMID: 17369853
-
DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells.J Mol Biol. 2001 Apr 13;307(5):1235-45. doi: 10.1006/jmbi.2001.4564. J Mol Biol. 2001. PMID: 11292338
-
Cancer therapy and replication stress: forks on the road to perdition.Adv Clin Chem. 2015;69:91-138. doi: 10.1016/bs.acc.2014.12.003. Epub 2015 Feb 7. Adv Clin Chem. 2015. PMID: 25934360 Review.
-
Targeting Replication Stress Response Pathways to Enhance Genotoxic Chemo- and Radiotherapy.Molecules. 2022 Jul 25;27(15):4736. doi: 10.3390/molecules27154736. Molecules. 2022. PMID: 35897913 Free PMC article. Review.
Cited by
-
The function of histone acetylation in cervical cancer development.Biosci Rep. 2019 Apr 12;39(4):BSR20190527. doi: 10.1042/BSR20190527. Print 2019 Apr 30. Biosci Rep. 2019. PMID: 30886064 Free PMC article. Review.
-
Autism-Associated Vigilin Depletion Impairs DNA Damage Repair.Mol Cell Biol. 2021 Jun 23;41(7):e0008221. doi: 10.1128/MCB.00082-21. Epub 2021 Jun 23. Mol Cell Biol. 2021. PMID: 33941620 Free PMC article.
-
Role of the Histone Acetyl Transferase MOF and the Histone Deacetylase Sirtuins in Regulation of H4K16ac During DNA Damage Repair and Metabolic Programming: Implications in Cancer and Aging.Subcell Biochem. 2022;100:115-141. doi: 10.1007/978-3-031-07634-3_4. Subcell Biochem. 2022. PMID: 36301493
-
Heat-induced SIRT1-mediated H4K16ac deacetylation impairs resection and SMARCAD1 recruitment to double strand breaks.iScience. 2022 Mar 23;25(4):104142. doi: 10.1016/j.isci.2022.104142. eCollection 2022 Apr 15. iScience. 2022. PMID: 35434547 Free PMC article.
-
Caspase-2 regulates S-phase cell cycle events to protect from DNA damage accumulation independent of apoptosis.Oncogene. 2022 Jan;41(2):204-219. doi: 10.1038/s41388-021-02085-w. Epub 2021 Oct 30. Oncogene. 2022. PMID: 34718349 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
