Cytomegaloviruses in a Community of Wild Nonhuman Primates in Taï National Park, Côte D'Ivoire
- PMID: 29286318
- PMCID: PMC5795424
- DOI: 10.3390/v10010011
Cytomegaloviruses in a Community of Wild Nonhuman Primates in Taï National Park, Côte D'Ivoire
Abstract
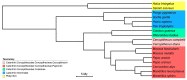
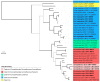
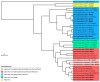
Cytomegaloviruses (CMVs) are known to infect many mammals, including a number of nonhuman primates (NHPs). However, most data available arose from studies led on captive individuals and little is known about CMV diversity in wild NHPs. Here, we analyzed a community of wild nonhuman primates (seven species) in Taï National Park (TNP), Côte d'Ivoire, with two PCR systems targeting betaherpesviruses. CMV DNA was detected in 17/87 primates (4/7 species). Six novel CMVs were identified in sooty mangabeys, Campbell's monkeys and Diana monkeys, respectively. In 3/17 positive individuals (from three NHP species), different CMVs were co-detected. A major part of the glycoprotein B coding sequences of the novel viruses was amplified and sequenced, and phylogenetic analyses were performed that included three previously discovered CMVs of western red colobus from TNP and published CMVs from other NHP species and geographic locations. We find that, despite this locally intensified sampling, NHP CMVs from TNP are completely host-specific, pinpointing the absence or rarity of cross-species transmission. We also show that on longer timescales the evolution of CMVs is characterized by frequent co-divergence with their hosts, although other processes, including lineage duplication and host switching, also have to be invoked to fully explain their evolutionary relationships.
Keywords: Côte d’Ivoire; Taï National Park; co-divergence; cytomegalovirus; genetic diversity; host specificity; nonhuman primate.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Genetic identification of cytomegaloviruses in a rural population of Côte d'Ivoire.Virol J. 2015 Oct 5;12:155. doi: 10.1186/s12985-015-0394-1. Virol J. 2015. PMID: 26437859 Free PMC article.
-
Prevalence and genetic diversity of simian immunodeficiency virus infection in wild-living red colobus monkeys (Piliocolobus badius badius) from the Taï forest, Côte d'Ivoire SIVwrc in wild-living western red colobus monkeys.Infect Genet Evol. 2008 Jan;8(1):1-14. doi: 10.1016/j.meegid.2007.08.004. Epub 2007 Sep 4. Infect Genet Evol. 2008. PMID: 17916449
-
DNA Polymerase Sequences of New World Monkey Cytomegaloviruses: Another Molecular Marker with Which To Infer Platyrrhini Systematics.J Virol. 2018 Aug 29;92(18):e00980-18. doi: 10.1128/JVI.00980-18. Print 2018 Sep 15. J Virol. 2018. PMID: 29976674 Free PMC article.
-
Evolution and Genetic Diversity of Primate Cytomegaloviruses.Microorganisms. 2020 Apr 25;8(5):624. doi: 10.3390/microorganisms8050624. Microorganisms. 2020. PMID: 32344906 Free PMC article. Review.
-
Rhesus monkeys for a nonhuman primate model of cytomegalovirus infections.Curr Opin Virol. 2017 Aug;25:126-133. doi: 10.1016/j.coviro.2017.08.005. Epub 2017 Sep 6. Curr Opin Virol. 2017. PMID: 28888133 Free PMC article. Review.
Cited by
-
Multiple DNA viruses identified in multimammate mouse (Mastomys natalensis) populations from across regions of sub-Saharan Africa.Arch Virol. 2020 Oct;165(10):2291-2299. doi: 10.1007/s00705-020-04738-9. Epub 2020 Aug 4. Arch Virol. 2020. PMID: 32754877 Free PMC article.
-
Cytomegalovirus distribution and evolution in hominines.Virus Evol. 2019 Aug 1;5(2):vez015. doi: 10.1093/ve/vez015. eCollection 2019 Jul. Virus Evol. 2019. PMID: 31384482 Free PMC article.
-
Molecular Detection and Genetic Diversity of Cytomegaloviruses and Lymphocryptoviruses in Free-Roaming and Captive African Green Monkeys (Chlorocebus sabaeus).Int J Mol Sci. 2024 Mar 14;25(6):3272. doi: 10.3390/ijms25063272. Int J Mol Sci. 2024. PMID: 38542246 Free PMC article.
-
Factors influencing bacterial microbiome composition in a wild non-human primate community in Taï National Park, Côte d'Ivoire.ISME J. 2018 Oct;12(10):2559-2574. doi: 10.1038/s41396-018-0166-1. Epub 2018 Jun 28. ISME J. 2018. PMID: 29955140 Free PMC article.
-
Viruses in saliva from sanctuary chimpanzees (Pan troglodytes) in Republic of Congo and Uganda.PLoS One. 2023 Jun 29;18(6):e0288007. doi: 10.1371/journal.pone.0288007. eCollection 2023. PLoS One. 2023. PMID: 37384730 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

