The inositol pyrophosphate pathway in health and diseases
- PMID: 29282838
- PMCID: PMC6383672
- DOI: 10.1111/brv.12392
The inositol pyrophosphate pathway in health and diseases
Abstract
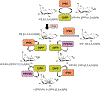
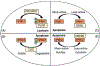
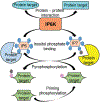
Inositol pyrophosphates (IPPs) are present in organisms ranging from plants, slime moulds and fungi to mammals. Distinct classes of kinases generate different forms of energetic diphosphate-containing IPPs from inositol phosphates (IPs). Conversely, polyphosphate phosphohydrolase enzymes dephosphorylate IPPs to regenerate the respective IPs. IPPs and/or their metabolizing enzymes regulate various cell biological processes by modulating many proteins via diverse mechanisms. In the last decade, extensive research has been conducted in mammalian systems, particularly in knockout mouse models of relevant enzymes. Results obtained from these studies suggest impacts of the IPP pathway on organ development, especially of brain and testis. Conversely, deletion of specific enzymes in the pathway protects mice from various diseases such as diet-induced obesity (DIO), type-2 diabetes (T2D), fatty liver, bacterial infection, thromboembolism, cancer metastasis and aging. Furthermore, pharmacological inhibition of the same class of enzymes in mice validates the therapeutic importance of this pathway in cardio-metabolic diseases. This review critically analyses these findings and summarizes the significance of the IPP pathway in mammalian health and diseases. It also evaluates benefits and risks of targeting this pathway in disease therapies. Finally, future directions of mammalian IPP research are discussed.
Keywords: DIPP; IP6K; PPIP5K; TNP; aging; cancer; cardiovascular disease; development; diabetes; inositol pyrophosphate; obesity.
© 2017 Cambridge Philosophical Society.
Figures


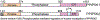

Similar articles
-
The inositol pyrophosphate metabolism of Dictyostelium discoideum does not regulate inorganic polyphosphate (polyP) synthesis.Adv Biol Regul. 2022 Jan;83:100835. doi: 10.1016/j.jbior.2021.100835. Epub 2021 Nov 10. Adv Biol Regul. 2022. PMID: 34782304 Free PMC article.
-
Inositol Pyrophosphates: Signaling Molecules with Pleiotropic Actions in Mammals.Molecules. 2020 May 8;25(9):2208. doi: 10.3390/molecules25092208. Molecules. 2020. PMID: 32397291 Free PMC article. Review.
-
Inositol pyrophosphates as mammalian cell signals.Sci Signal. 2011 Aug 23;4(188):re1. doi: 10.1126/scisignal.2001958. Sci Signal. 2011. PMID: 21878680 Free PMC article. Review.
-
TNP [N2-(m-Trifluorobenzyl), N6-(p-nitrobenzyl)purine] ameliorates diet induced obesity and insulin resistance via inhibition of the IP6K1 pathway.Mol Metab. 2016 Aug 21;5(10):903-917. doi: 10.1016/j.molmet.2016.08.008. eCollection 2016 Oct. Mol Metab. 2016. PMID: 27689003 Free PMC article.
-
VIH2 Regulates the Synthesis of Inositol Pyrophosphate InsP8 and Jasmonate-Dependent Defenses in Arabidopsis.Plant Cell. 2015 Apr;27(4):1082-97. doi: 10.1105/tpc.114.135160. Epub 2015 Apr 21. Plant Cell. 2015. PMID: 25901085 Free PMC article.
Cited by
-
Metabolic supervision by PPIP5K, an inositol pyrophosphate kinase/phosphatase, controls proliferation of the HCT116 tumor cell line.Proc Natl Acad Sci U S A. 2021 Mar 9;118(10):e2020187118. doi: 10.1073/pnas.2020187118. Proc Natl Acad Sci U S A. 2021. PMID: 33649228 Free PMC article.
-
Physiologically Active Molecules and Functional Properties of Soybeans in Human Health-A Current Perspective.Int J Mol Sci. 2021 Apr 14;22(8):4054. doi: 10.3390/ijms22084054. Int J Mol Sci. 2021. PMID: 33920015 Free PMC article. Review.
-
Fragment-Based Screening Identifies New Quinazolinone-Based Inositol Hexakisphosphate Kinase (IP6K) Inhibitors.ACS Med Chem Lett. 2023 Nov 28;14(12):1760-1766. doi: 10.1021/acsmedchemlett.3c00409. eCollection 2023 Dec 14. ACS Med Chem Lett. 2023. PMID: 38116421 Free PMC article.
-
A two-way switch for inositol pyrophosphate signaling: Evolutionary history and biological significance of a unique, bifunctional kinase/phosphatase.Adv Biol Regul. 2020 Jan;75:100674. doi: 10.1016/j.jbior.2019.100674. Epub 2019 Nov 14. Adv Biol Regul. 2020. PMID: 31776069 Free PMC article. Review.
-
Inositol hexakisphosphate kinase 3 promotes focal adhesion turnover via interactions with dynein intermediate chain 2.Proc Natl Acad Sci U S A. 2019 Feb 19;116(8):3278-3287. doi: 10.1073/pnas.1817001116. Epub 2019 Feb 4. Proc Natl Acad Sci U S A. 2019. PMID: 30718399 Free PMC article.
References
-
- AUESUKAREE C, TOCHIO H, SHIRAKAWA M, KANEKO Y & HARASHIMA S (2005). Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. Journal of Biological Chemistry 280, 25127–25133. - PubMed
-
- AZEVEDO C & SAIARDI A (2006). Extraction and analysis of soluble inositol polyphosphates from yeast. Nature Protocols 1, 2416–2422. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

