Detection and full genome characterization of two beta CoV viruses related to Middle East respiratory syndrome from bats in Italy
- PMID: 29258555
- PMCID: PMC5735805
- DOI: 10.1186/s12985-017-0907-1
Detection and full genome characterization of two beta CoV viruses related to Middle East respiratory syndrome from bats in Italy
Erratum in
-
Correction to: Detection and full genome characterization of two beta CoV viruses related to Middle East respiratory syndrome from bats in Italy.Virol J. 2018 Jan 12;15(1):10. doi: 10.1186/s12985-018-0921-y. Virol J. 2018. PMID: 29329554 Free PMC article.
Abstract
Background: Middle East respiratory syndrome coronavirus (MERS-CoV), which belongs to beta group of coronavirus, can infect multiple host species and causes severe diseases in humans. Multiple surveillance and phylogenetic studies suggest a bat origin. In this study, we describe the detection and full genome characterization of two CoVs closely related to MERS-CoV from two Italian bats, Pipistrellus kuhlii and Hypsugo savii.
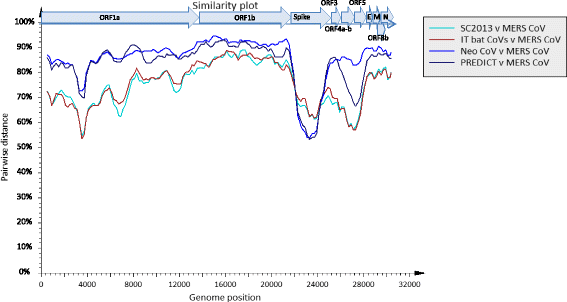
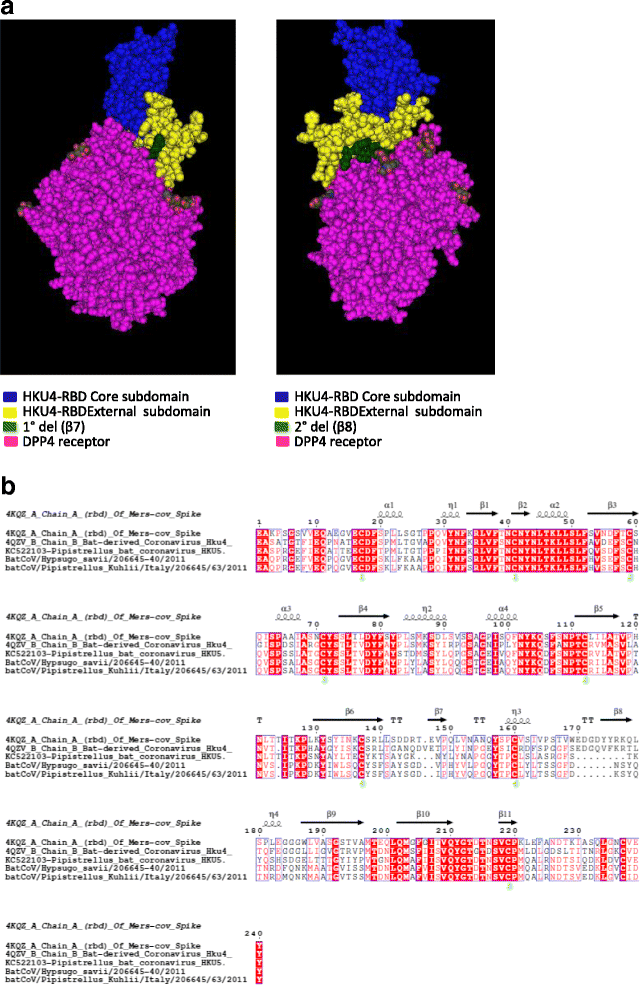
Methods: Pool of viscera were tested by a pan-coronavirus RT-PCR. Virus isolation was attempted by inoculation in different cell lines. Full genome sequencing was performed using the Ion Torrent platform and phylogenetic trees were performed using IQtree software. Similarity plots of CoV clade c genomes were generated by using SSE v1.2. The three dimensional macromolecular structure (3DMMS) of the receptor binding domain (RBD) in the S protein was predicted by sequence-homology method using the protein data bank (PDB).
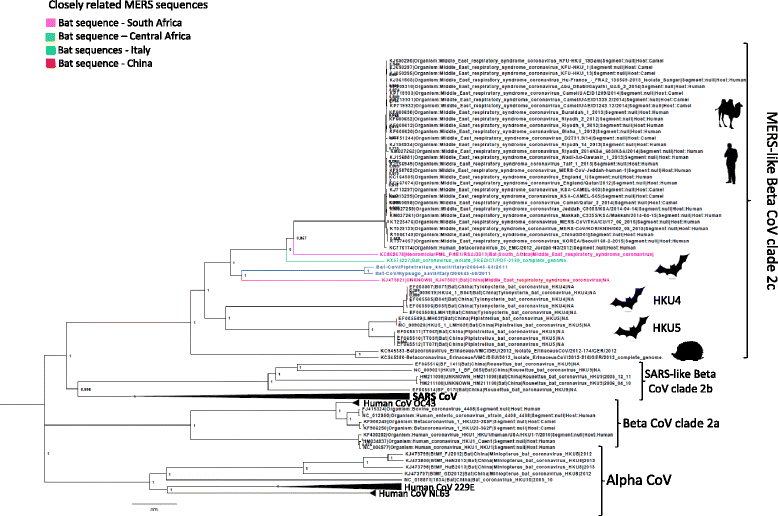
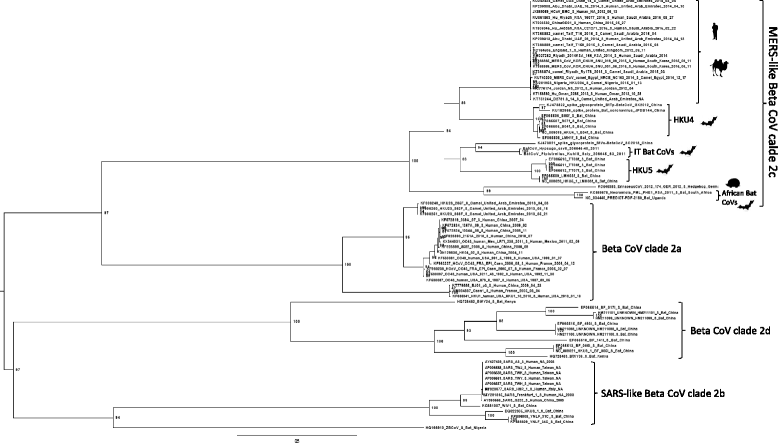
Results: Both samples resulted positive to the pan-coronavirus RT-PCR (IT-batCoVs) and their genome organization showed identical pattern of MERS CoV. Phylogenetic analysis showed a monophyletic group placed in the Beta2c clade formed by MERS-CoV sequences originating from humans and camels and bat-related sequences from Africa, Italy and China. The comparison of the secondary and 3DMMS of the RBD of IT-batCoVs with MERS, HKU4 and HKU5 bat sequences showed two aa deletions located in a region corresponding to the external subdomain of MERS-RBD in IT-batCoV and HKU5 RBDs.
Conclusions: This study reported two beta CoVs closely related to MERS that were obtained from two bats belonging to two commonly recorded species in Italy (P. kuhlii and H. savii). The analysis of the RBD showed similar structure in IT-batCoVs and HKU5 respect to HKU4 sequences. Since the RBD domain of HKU4 but not HKU5 can bind to the human DPP4 receptor for MERS-CoV, it is possible to suggest also for IT-batCoVs the absence of DPP4-binding potential. More surveillance studies are needed to better investigate the potential intermediate hosts that may play a role in the interspecies transmission of known and currently unknown coronaviruses with particular attention to the S protein and the receptor specificity and binding affinity.
Keywords: Bats; Full genome sequencing; Italy; MERS-like Beta-CoV viruses; Phylogenetic and molecular analyses.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Rapid detection of MERS coronavirus-like viruses in bats: pote1ntial for tracking MERS coronavirus transmission and animal origin.Emerg Microbes Infect. 2018 Mar 7;7(1):18. doi: 10.1038/s41426-017-0016-7. Emerg Microbes Infect. 2018. PMID: 29511173 Free PMC article.
-
Further Evidence for Bats as the Evolutionary Source of Middle East Respiratory Syndrome Coronavirus.mBio. 2017 Apr 4;8(2):e00373-17. doi: 10.1128/mBio.00373-17. mBio. 2017. PMID: 28377531 Free PMC article.
-
Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26.Cell Host Microbe. 2014 Sep 10;16(3):328-37. doi: 10.1016/j.chom.2014.08.009. Cell Host Microbe. 2014. PMID: 25211075 Free PMC article.
-
Middle East respiratory syndrome coronavirus (MERS-CoV): animal to human interaction.Pathog Glob Health. 2015;109(8):354-62. doi: 10.1080/20477724.2015.1122852. Pathog Glob Health. 2015. PMID: 26924345 Free PMC article. Review.
-
[A novel coronavirus, MERS-CoV].Uirusu. 2013;63(1):1-6. doi: 10.2222/jsv.63.1. Uirusu. 2013. PMID: 24769571 Review. Japanese.
Cited by
-
Global Epidemiology of Bat Coronaviruses.Viruses. 2019 Feb 20;11(2):174. doi: 10.3390/v11020174. Viruses. 2019. PMID: 30791586 Free PMC article. Review.
-
Presence of Alphacoronavirus in Tree- and Crevice-Dwelling Bats from Portugal.Viruses. 2024 Mar 12;16(3):434. doi: 10.3390/v16030434. Viruses. 2024. PMID: 38543799 Free PMC article.
-
Novel Insights Into Immune Systems of Bats.Front Immunol. 2020 Jan 24;11:26. doi: 10.3389/fimmu.2020.00026. eCollection 2020. Front Immunol. 2020. PMID: 32117225 Free PMC article. Review.
-
Single cell atlas for 11 non-model mammals, reptiles and birds.Nat Commun. 2021 Dec 6;12(1):7083. doi: 10.1038/s41467-021-27162-2. Nat Commun. 2021. PMID: 34873160 Free PMC article.
-
Evolutionary trajectory of SARS-CoV-2 and emerging variants.Virol J. 2021 Aug 13;18(1):166. doi: 10.1186/s12985-021-01633-w. Virol J. 2021. PMID: 34389034 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

