Overexpression of OsCIPK30 Enhances Plant Tolerance to Rice stripe virus
- PMID: 29225594
- PMCID: PMC5705616
- DOI: 10.3389/fmicb.2017.02322
Overexpression of OsCIPK30 Enhances Plant Tolerance to Rice stripe virus
Abstract
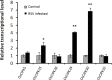
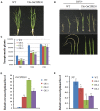
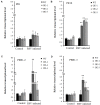
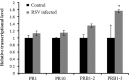
Rice stripe virus (RSV) causes a severe disease in Oryza sativa (rice) in many Eastern Asian countries. The NS3 protein of RSV is a viral suppressor of RNA silencing, but plant host factors interacting with NS3 have not been reported yet. Here, we present evidence that expression of RSV NS3 in Arabidopsis thaliana causes developmental abnormalities. Through yeast two-hybrid screening and a luciferase complementation imaging assay, we demonstrate that RSV NS3 interacted with OsCIPK30, a CBL (calcineurin B-like proteins)-interaction protein kinase protein. Furthermore, OsCIPK30 was overexpressed to investigate the function of OsCIPK30 in rice. Our investigation showed that overexpression of OsCIPK30 in rice could delay the RSV symptoms and show milder RSV symptoms. In addition, the expression of pathogenesis-related genes was increased in OsCIPK30 transgenic rice. These results suggest that overexpression of OsCIPK30 positively regulates pathogenesis-related genes to enhance the tolerance to RSV in rice. Our findings provide new insight into the molecular mechanism underlying resistance to RSV disease.
Keywords: NS3; OsCIPK30; Rice stripe virus; interaction; pathogenesis-related genes.
Figures



Similar articles
-
Interaction between Rice stripe virus disease-specific protein and host PsbP enhances virus symptoms.Mol Plant. 2014 Apr;7(4):691-708. doi: 10.1093/mp/sst158. Epub 2013 Nov 8. Mol Plant. 2014. PMID: 24214893
-
Characterization and subcellular localization of an RNA silencing suppressor encoded by Rice stripe tenuivirus.Virology. 2009 Apr 25;387(1):29-40. doi: 10.1016/j.virol.2009.01.045. Epub 2009 Feb 28. Virology. 2009. PMID: 19251298
-
Rice stripe virus NS3 protein regulates primary miRNA processing through association with the miRNA biogenesis factor OsDRB1 and facilitates virus infection in rice.PLoS Pathog. 2017 Oct 4;13(10):e1006662. doi: 10.1371/journal.ppat.1006662. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 28977024 Free PMC article.
-
Identification of residues or motif(s) of the rice stripe virus NS3 protein required for self-interaction and for silencing suppressor activity.Virus Res. 2017 May 2;235:14-23. doi: 10.1016/j.virusres.2017.03.022. Epub 2017 Apr 6. Virus Res. 2017. PMID: 28392445
-
Current Insights into Research on Rice stripe virus.Plant Pathol J. 2013 Sep;29(3):223-33. doi: 10.5423/PPJ.RW.10.2012.0158. Plant Pathol J. 2013. PMID: 25288949 Free PMC article. Review.
Cited by
-
Exploring the Diversity of Mechanisms Associated With Plant Tolerance to Virus Infection.Front Plant Sci. 2018 Nov 2;9:1575. doi: 10.3389/fpls.2018.01575. eCollection 2018. Front Plant Sci. 2018. PMID: 30450108 Free PMC article. Review.
-
Potato calcineurin B-like protein CBL4, interacting with calcineurin B-like protein-interacting protein kinase CIPK2, positively regulates plant resistance to stem canker caused by Rhizoctonia solani.Front Microbiol. 2023 Jan 4;13:1032900. doi: 10.3389/fmicb.2022.1032900. eCollection 2022. Front Microbiol. 2023. PMID: 36687567 Free PMC article.
-
Engineering plant immune circuit: walking to the bright future with a novel toolbox.Plant Biotechnol J. 2023 Jan;21(1):17-45. doi: 10.1111/pbi.13916. Epub 2022 Sep 27. Plant Biotechnol J. 2023. PMID: 36036862 Free PMC article. Review.
-
Temporal expression of defence and susceptibility genes and tospovirus accumulation in capsicum chlorosis virus-infected capsicum.Arch Virol. 2022 Apr;167(4):1061-1074. doi: 10.1007/s00705-022-05401-1. Epub 2022 Mar 4. Arch Virol. 2022. PMID: 35246732 Free PMC article.
-
Pathogenesis-related protein 10 in resistance to biotic stress: progress in elucidating functions, regulation and modes of action.Front Plant Sci. 2023 Jul 4;14:1193873. doi: 10.3389/fpls.2023.1193873. eCollection 2023. Front Plant Sci. 2023. PMID: 37469770 Free PMC article.
References
-
- Applied Biosystems (1997). “Relative quantitation of gene expression” in User Bulletin #2: ABI Prism 7700 Sequence Detection System, ed. Livak K. J. (Foster City, CA: Applied Biosystems; ).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

