A Unique ISR Program Determines Cellular Responses to Chronic Stress
- PMID: 29220654
- PMCID: PMC5730339
- DOI: 10.1016/j.molcel.2017.11.007
A Unique ISR Program Determines Cellular Responses to Chronic Stress
Abstract
The integrated stress response (ISR) is a homeostatic mechanism induced by endoplasmic reticulum (ER) stress. In acute/transient ER stress, decreased global protein synthesis and increased uORF mRNA translation are followed by normalization of protein synthesis. Here, we report a dramatically different response during chronic ER stress. This chronic ISR program is characterized by persistently elevated uORF mRNA translation and concurrent gene expression reprogramming, which permits simultaneous stress sensing and proteostasis. The program includes PERK-dependent switching to an eIF3-dependent translation initiation mechanism, resulting in partial, but not complete, translational recovery, which, together with transcriptional reprogramming, selectively bolsters expression of proteins with ER functions. Coordination of transcriptional and translational reprogramming prevents ER dysfunction and inhibits "foamy cell" development, thus establishing a molecular basis for understanding human diseases associated with ER dysfunction.
Keywords: ER stress; PERK; eIF2; eIF2B; eIF3; integrated stress response; mRNA translation; protein synthesis; stress signaling; unfolded protein response.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


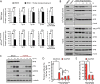


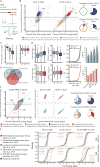
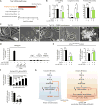
Similar articles
-
Methods for analyzing eIF2 kinases and translational control in the unfolded protein response.Methods Enzymol. 2011;490:333-56. doi: 10.1016/B978-0-12-385114-7.00019-2. Methods Enzymol. 2011. PMID: 21266259
-
Mammalian ECD Protein Is a Novel Negative Regulator of the PERK Arm of the Unfolded Protein Response.Mol Cell Biol. 2017 Aug 28;37(18):e00030-17. doi: 10.1128/MCB.00030-17. Print 2017 Sep 15. Mol Cell Biol. 2017. PMID: 28652267 Free PMC article.
-
Effects of muscular dystrophy, exercise and blocking activin receptor IIB ligands on the unfolded protein response and oxidative stress.Free Radic Biol Med. 2016 Oct;99:308-322. doi: 10.1016/j.freeradbiomed.2016.08.017. Epub 2016 Aug 20. Free Radic Biol Med. 2016. PMID: 27554968
-
Mechanisms of disordered neurodegenerative function: concepts and facts about the different roles of the protein kinase RNA-like endoplasmic reticulum kinase (PERK).Rev Neurosci. 2018 Jun 27;29(4):387-415. doi: 10.1515/revneuro-2017-0071. Rev Neurosci. 2018. PMID: 29303785 Review.
-
[Endoplasmic reticulum-mediated integrated stress response].Sheng Li Ke Xue Jin Zhan. 2013 Aug;44(4):241-6. Sheng Li Ke Xue Jin Zhan. 2013. PMID: 24228513 Review. Chinese.
Cited by
-
SMYD5 methylation of rpL40 links ribosomal output to gastric cancer.Nature. 2024 Aug;632(8025):656-663. doi: 10.1038/s41586-024-07718-0. Epub 2024 Jul 24. Nature. 2024. PMID: 39048817 Free PMC article.
-
RITA requires eIF2α-dependent modulation of mRNA translation for its anti-cancer activity.Cell Death Dis. 2019 Nov 7;10(11):845. doi: 10.1038/s41419-019-2074-3. Cell Death Dis. 2019. PMID: 31699971 Free PMC article.
-
Alternative mechanisms of translation initiation: An emerging dynamic regulator of the proteome in health and disease.Life Sci. 2018 Nov 1;212:138-144. doi: 10.1016/j.lfs.2018.09.054. Epub 2018 Oct 2. Life Sci. 2018. PMID: 30290184 Free PMC article. Review.
-
Simultaneous measurement of nascent transcriptome and translatome using 4-thiouridine metabolic RNA labeling and translating ribosome affinity purification.Nucleic Acids Res. 2023 Aug 11;51(14):e76. doi: 10.1093/nar/gkad545. Nucleic Acids Res. 2023. PMID: 37378452 Free PMC article.
-
Retrograde signaling by a mtDNA-encoded non-coding RNA preserves mitochondrial bioenergetics.Commun Biol. 2020 Oct 30;3(1):626. doi: 10.1038/s42003-020-01322-4. Commun Biol. 2020. PMID: 33127975 Free PMC article.
References
-
- Atkins C, Liu Q, Minthorn E, Zhang SY, Figueroa DJ, Moss K, Stanley TB, Sanders B, Goetz A, Gaul N, et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer research. 2013;73:1993–2002. - PubMed
-
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nature chemical biology. 2006;2:213–220. - PubMed
-
- Castello A, Horos R, Strein C, Fischer B, Eichelbaum K, Steinmetz LM, Krijgsveld J, Hentze MW. Comprehensive Identification of RNA-Binding Proteins by RNA Interactome Capture. Methods in molecular biology (Clifton, N.J.) 2016;1358:131–139. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

