Single-molecule studies contrast ordered DNA replication with stochastic translesion synthesis
- PMID: 29210356
- PMCID: PMC5731819
- DOI: 10.7554/eLife.32177
Single-molecule studies contrast ordered DNA replication with stochastic translesion synthesis
Abstract
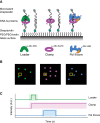
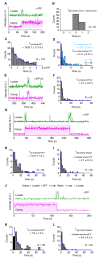
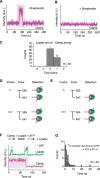
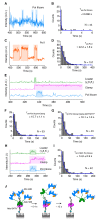
High fidelity replicative DNA polymerases are unable to synthesize past DNA adducts that result from diverse chemicals, reactive oxygen species or UV light. To bypass these replication blocks, cells utilize specialized translesion DNA polymerases that are intrinsically error prone and associated with mutagenesis, drug resistance, and cancer. How untimely access of translesion polymerases to DNA is prevented is poorly understood. Here we use co-localization single-molecule spectroscopy (CoSMoS) to follow the exchange of the E. coli replicative DNA polymerase Pol IIIcore with the translesion polymerases Pol II and Pol IV. We find that in contrast to the toolbelt model, the replicative and translesion polymerases do not form a stable complex on one clamp but alternate their binding. Furthermore, while the loading of clamp and Pol IIIcore is highly organized, the exchange with the translesion polymerases is stochastic and is not determined by lesion-recognition but instead a concentration-dependent competition between the polymerases.
Keywords: CoSMoS; DNA replication; E. coli; biochemistry; biophysics; single-molecule; structural biology; translesion DNA synthesis.
Conflict of interest statement
No competing interests declared.
Figures



Similar articles
-
Translesion synthesis in Escherichia coli: lessons from the NarI mutation hot spot.DNA Repair (Amst). 2007 Jul 1;6(7):1032-41. doi: 10.1016/j.dnarep.2007.02.021. Epub 2007 Apr 2. DNA Repair (Amst). 2007. PMID: 17403618 Review.
-
A Comprehensive View of Translesion Synthesis in Escherichia coli.Microbiol Mol Biol Rev. 2020 Jun 17;84(3):e00002-20. doi: 10.1128/MMBR.00002-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32554755 Free PMC article. Review.
-
The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo.EMBO Rep. 2002 Jan;3(1):45-9. doi: 10.1093/embo-reports/kvf007. Epub 2001 Dec 19. EMBO Rep. 2002. PMID: 11751576 Free PMC article.
-
Exchange between Escherichia coli polymerases II and III on a processivity clamp.Nucleic Acids Res. 2016 Feb 29;44(4):1681-90. doi: 10.1093/nar/gkv1375. Epub 2015 Dec 10. Nucleic Acids Res. 2016. PMID: 26657641 Free PMC article.
-
Polymerase exchange on single DNA molecules reveals processivity clamp control of translesion synthesis.Proc Natl Acad Sci U S A. 2014 May 27;111(21):7647-52. doi: 10.1073/pnas.1321076111. Epub 2014 May 13. Proc Natl Acad Sci U S A. 2014. PMID: 24825884 Free PMC article.
Cited by
-
Escherichia coli DNA replication: the old model organism still holds many surprises.FEMS Microbiol Rev. 2024 Jun 20;48(4):fuae018. doi: 10.1093/femsre/fuae018. FEMS Microbiol Rev. 2024. PMID: 38982189 Free PMC article. Review.
-
Frequent template switching in postreplication gaps: suppression of deleterious consequences by the Escherichia coli Uup and RadD proteins.Nucleic Acids Res. 2020 Jan 10;48(1):212-230. doi: 10.1093/nar/gkz960. Nucleic Acids Res. 2020. PMID: 31665437 Free PMC article.
-
Translesion Synthesis or Repair by Specialized DNA Polymerases Limits Excessive Genomic Instability upon Replication Stress.Int J Mol Sci. 2021 Apr 10;22(8):3924. doi: 10.3390/ijms22083924. Int J Mol Sci. 2021. PMID: 33920223 Free PMC article. Review.
-
The SOS system: A complex and tightly regulated response to DNA damage.Environ Mol Mutagen. 2019 May;60(4):368-384. doi: 10.1002/em.22267. Epub 2019 Jan 7. Environ Mol Mutagen. 2019. PMID: 30447030 Free PMC article. Review.
-
Dynamics of Proteins and Macromolecular Machines in Escherichia coli.EcoSal Plus. 2021 Dec 15;9(2):eESP00112020. doi: 10.1128/ecosalplus.ESP-0011-2020. Epub 2021 Jun 1. EcoSal Plus. 2021. PMID: 34060908 Free PMC article. Review.
References
-
- Bonner CA, Randall SK, Rayssiguier C, Radman M, Eritja R, Kaplan BE, McEntee K, Goodman MF. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. The Journal of Biological Chemistry. 1988;263:18946–18952. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

