Drug-Drug Interactions between Atorvastatin and Dronedarone Mediated by Monomeric CYP3A4
- PMID: 29200287
- PMCID: PMC5800941
- DOI: 10.1021/acs.biochem.7b01012
Drug-Drug Interactions between Atorvastatin and Dronedarone Mediated by Monomeric CYP3A4
Abstract
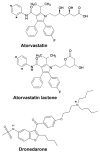
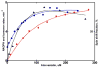
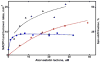
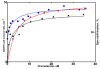
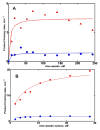
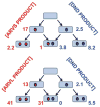
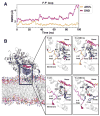
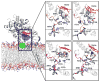
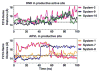
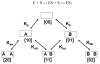
Heterotropic interactions between atorvastatin (ARVS) and dronedarone (DND) have been deciphered using global analysis of the results of binding and turnover experiments for pure drugs and their mixtures. The in vivo presence of atorvastatin lactone (ARVL) was explicitly taken into account by using pure ARVL in analogous experiments. Both ARVL and ARVS inhibit DND binding and metabolism, while a significantly higher affinity of CYP3A4 for ARVL makes the latter the main modulator of activity (effector) in this system. Molecular dynamics simulations reveal significantly different modes of interactions of DND and ARVL with the substrate binding pocket and with a peripheral allosteric site. Interactions of both substrates with residues F213 and F219 at the allosteric site play a critical role in the communication of conformational changes induced by effector binding to productive binding of the substrate at the catalytic site.
Figures


Similar articles
-
Midazolam as a Probe for Drug-Drug Interactions Mediated by CYP3A4: Homotropic Allosteric Mechanism of Site-Specific Hydroxylation.Biochemistry. 2021 Jun 1;60(21):1670-1681. doi: 10.1021/acs.biochem.1c00161. Epub 2021 May 20. Biochemistry. 2021. PMID: 34015213 Free PMC article.
-
Mechanism of drug-drug interactions mediated by human cytochrome P450 CYP3A4 monomer.Biochemistry. 2015 Apr 7;54(13):2227-39. doi: 10.1021/acs.biochem.5b00079. Epub 2015 Mar 25. Biochemistry. 2015. PMID: 25777547 Free PMC article.
-
Inactivation of Human Cytochrome P450 3A4 and 3A5 by Dronedarone and N-Desbutyl Dronedarone.Mol Pharmacol. 2016 Jan;89(1):1-13. doi: 10.1124/mol.115.100891. Epub 2015 Oct 21. Mol Pharmacol. 2016. PMID: 26490246
-
Challenges in assignment of allosteric effects in cytochrome P450-catalyzed substrate oxidations to structural dynamics in the hemoprotein architecture.J Inorg Biochem. 2017 Feb;167:100-115. doi: 10.1016/j.jinorgbio.2016.11.025. Epub 2016 Nov 25. J Inorg Biochem. 2017. PMID: 27919007 Review.
-
Protein Allostery and Conformational Dynamics.Chem Rev. 2016 Jun 8;116(11):6503-15. doi: 10.1021/acs.chemrev.5b00590. Epub 2016 Feb 15. Chem Rev. 2016. PMID: 26876046 Free PMC article. Review.
Cited by
-
Midazolam as a Probe for Drug-Drug Interactions Mediated by CYP3A4: Homotropic Allosteric Mechanism of Site-Specific Hydroxylation.Biochemistry. 2021 Jun 1;60(21):1670-1681. doi: 10.1021/acs.biochem.1c00161. Epub 2021 May 20. Biochemistry. 2021. PMID: 34015213 Free PMC article.
-
Allosteric Interactions in Human Cytochrome P450 CYP3A4: The Role of Phenylalanine 213.Biochemistry. 2019 Mar 12;58(10):1411-1422. doi: 10.1021/acs.biochem.8b01268. Epub 2019 Feb 28. Biochemistry. 2019. PMID: 30785734 Free PMC article.
-
Dynamics and Location of the Allosteric Midazolam Site in Cytochrome P4503A4 in Lipid Nanodiscs.Biochemistry. 2020 Feb 18;59(6):766-779. doi: 10.1021/acs.biochem.9b01001. Epub 2020 Jan 27. Biochemistry. 2020. PMID: 31961139 Free PMC article.
-
Correction to "Allosteric Interactions in Human Cytochrome P450 CYP3A4: The Role of Phenylalanine 213".Biochemistry. 2019 Jun 18;58(24):2796. doi: 10.1021/acs.biochem.9b00438. Epub 2019 Jun 6. Biochemistry. 2019. PMID: 31181900 Free PMC article. No abstract available.
-
Steroid bioconjugation to a CYP3A4 allosteric site and its effect on substrate binding and coupling efficiency.Arch Biochem Biophys. 2018 Sep 1;653:90-96. doi: 10.1016/j.abb.2018.06.014. Epub 2018 Jun 26. Arch Biochem Biophys. 2018. PMID: 29958895 Free PMC article.
References
-
- Egnell A-C, Houston JB, Boyer CS. Predictive models of CYP3A4 heteroactivation: in vitro-in vivo scaling and pharmacophore modeling. Journal of Pharmacology and Experimental Therapeutics. 2005;312:926–937. - PubMed
-
- Mosa A, Neunzig J, Gerber A, Zapp J, Hannemann F, Pilak P, Bernhardt R. 2beta- and 16beta-hydroxylase activity of CYP11A1 and direct stimulatory effect of estrogens on pregnenolone formation. J Steroid Biochem Mol Biol 2015 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

