Serial femtosecond crystallography at the SACLA: breakthrough to dynamic structural biology
- PMID: 29196935
- PMCID: PMC5899704
- DOI: 10.1007/s12551-017-0344-9
Serial femtosecond crystallography at the SACLA: breakthrough to dynamic structural biology
Abstract
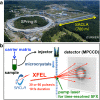
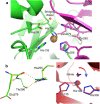
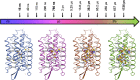
X-ray crystallography visualizes the world at the atomic level. It has been used as the most powerful technique for observing the three-dimensional structures of biological macromolecules and has pioneered structural biology. To determine a crystal structure with high resolution, it was traditionally required to prepare large crystals (> 200 μm). Later, synchrotron radiation facilities, such as SPring-8, that produce powerful X-rays were built. They enabled users to obtain good quality X-ray diffraction images even with smaller crystals (ca. 200-50 μm). In recent years, one of the most important technological innovations in structural biology has been the development of X-ray free electron lasers (XFELs). The SPring-8 Angstrom Compact free electron LAser (SACLA) in Japan generates the XFEL beam by accelerating electrons to relativistic speeds and directing them through in-vacuum, short-period undulators. Since user operation started in 2012, we have been involved in the development of serial femtosecond crystallography (SFX) measurement systems using XFEL at the SACLA. The SACLA generates X-rays a billion times brighter than SPring-8. The extremely bright XFEL pulses enable data collection with microcrystals (ca. 50-1 μm). Although many molecular analysis techniques exist, SFX is the only technique that can visualize radiation-damage-free structures of biological macromolecules at room temperature in atomic resolution and fast time resolution. Here, we review the achievements of the SACLA-SFX Project in the past 5 years. In particular, we focus on: (1) the measurement system for SFX; (2) experimental phasing by SFX; (3) enzyme chemistry based on damage-free room-temperature structures; and (4) molecular movie taken by time-resolved SFX.
Keywords: Bioinorganic chemistry; De novo phasing; Detergent; Membrane protein; Structure–function relationship; Time-resolved analysis.
Conflict of interest statement
Conflict of interest
Eiichi Mizohata declares that he has no conflict of interest. Takanori Nakane declares that he has no conflict of interest. Yohta Fukuda declares that he has no conflict of interest. Eriko Nango declares that she has no conflict of interest. So Iwata declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures
Similar articles
-
[What Kind of Measurements Can Be Made with an X-ray Free Electron Laser at SACLA?].Yakugaku Zasshi. 2022;142(5):479-485. doi: 10.1248/yakushi.21-00203-1. Yakugaku Zasshi. 2022. PMID: 35491153 Review. Japanese.
-
Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography.IUCrJ. 2017 Aug 8;4(Pt 5):639-647. doi: 10.1107/S2052252517008557. eCollection 2017 Sep 1. IUCrJ. 2017. PMID: 28989719 Free PMC article.
-
Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent.Proc Natl Acad Sci U S A. 2016 Nov 15;113(46):13039-13044. doi: 10.1073/pnas.1602531113. Epub 2016 Oct 31. Proc Natl Acad Sci U S A. 2016. PMID: 27799539 Free PMC article.
-
A Bright Future for Serial Femtosecond Crystallography with XFELs.Trends Biochem Sci. 2017 Sep;42(9):749-762. doi: 10.1016/j.tibs.2017.06.007. Epub 2017 Jul 18. Trends Biochem Sci. 2017. PMID: 28733116 Free PMC article. Review.
-
Dynamic Structural Biology Experiments at XFEL or Synchrotron Sources.Methods Mol Biol. 2021;2305:203-228. doi: 10.1007/978-1-0716-1406-8_11. Methods Mol Biol. 2021. PMID: 33950392 Review.
Cited by
-
Crystal structure of CmABCB1 multi-drug exporter in lipidic mesophase revealed by LCP-SFX.IUCrJ. 2021 Dec 23;9(Pt 1):134-145. doi: 10.1107/S2052252521011611. eCollection 2022 Jan 1. IUCrJ. 2021. PMID: 35059217 Free PMC article.
-
Foreword to 'Multiscale structural biology: biophysical principles and mechanisms underlying the action of bio-nanomachines', a special issue in Honour of Fumio Arisaka's 70th birthday.Biophys Rev. 2018 Apr;10(2):105-129. doi: 10.1007/s12551-018-0401-z. Epub 2018 Mar 2. Biophys Rev. 2018. PMID: 29500796 Free PMC article.
-
Experimental Protein Molecular Dynamics: Broadband Dielectric Spectroscopy coupled with nanoconfinement.Sci Rep. 2019 Nov 29;9(1):17988. doi: 10.1038/s41598-019-54562-8. Sci Rep. 2019. PMID: 31784681 Free PMC article.
-
In situ protein micro-crystal fabrication by cryo-FIB for electron diffraction.Biophys Rep. 2018;4(6):339-347. doi: 10.1007/s41048-018-0075-x. Epub 2018 Nov 14. Biophys Rep. 2018. PMID: 30596142 Free PMC article.
-
High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.IUCrJ. 2019 Apr 5;6(Pt 3):412-425. doi: 10.1107/S205225251900263X. eCollection 2019 May 1. IUCrJ. 2019. PMID: 31098022 Free PMC article.
References
-
- Antonyuk SV, Strange RW, Sawers G, Eady RR, Hasnain SS. Atomic resolution structures of resting-state, substrate- and product-complexed Cu-nitrite reductase provide insight into catalytic mechanism. Proc Natl Acad Sci U S A. 2005;102(34):12041–12046. doi: 10.1073/pnas.0504207102. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

