An activity-dependent proximity ligation platform for spatially resolved quantification of active enzymes in single cells
- PMID: 29176560
- PMCID: PMC5701173
- DOI: 10.1038/s41467-017-01854-0
An activity-dependent proximity ligation platform for spatially resolved quantification of active enzymes in single cells
Abstract
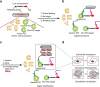
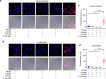
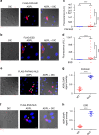
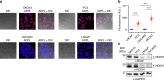
Integration of chemical probes into proteomic workflows enables the interrogation of protein activity, rather than abundance. Current methods limit the biological contexts that can be addressed due to sample homogenization, signal-averaging, and bias toward abundant proteins. Here we report a platform that integrates family-wide chemical probes with proximity-dependent oligonucleotide amplification and imaging to quantify enzyme activity in native contexts with high spatial resolution. Application of this method, activity-dependent proximity ligation (ADPL), to serine hydrolase and cysteine protease enzymes enables quantification of differential enzyme activity resulting from endogenous changes in localization and expression. In a competitive format, small-molecule target engagement with endogenous proteins in live cells can be quantified. Finally, retention of sample architecture enables interrogation of complex environments such as cellular co-culture and patient samples. ADPL should be amenable to diverse probe and protein families to detect active enzymes at scale and resolution out of reach with current methods.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Chemical probes for spatially resolved measurement of active enzymes in single cells.Methods Enzymol. 2019;628:243-262. doi: 10.1016/bs.mie.2019.06.017. Epub 2019 Jul 12. Methods Enzymol. 2019. PMID: 31668232 Free PMC article.
-
Ultrasensitive, multiplexed chemoproteomic profiling with soluble activity-dependent proximity ligation.Proc Natl Acad Sci U S A. 2019 Oct 22;116(43):21493-21500. doi: 10.1073/pnas.1912934116. Epub 2019 Oct 7. Proc Natl Acad Sci U S A. 2019. PMID: 31591248 Free PMC article.
-
Application of activity-based protein profiling to study enzyme function in adipocytes.Methods Enzymol. 2014;538:151-69. doi: 10.1016/B978-0-12-800280-3.00009-8. Methods Enzymol. 2014. PMID: 24529438 Free PMC article.
-
Strategies for detection and quantification of cysteine cathepsins-evolution from bench to bedside.Biochimie. 2016 Mar;122:48-61. doi: 10.1016/j.biochi.2015.07.029. Epub 2015 Aug 4. Biochimie. 2016. PMID: 26253694 Review.
-
Proteomic techniques and activity-based probes for the system-wide study of proteolysis.Biochimie. 2010 Nov;92(11):1705-14. doi: 10.1016/j.biochi.2010.04.027. Epub 2010 May 20. Biochimie. 2010. PMID: 20493233 Review.
Cited by
-
Increased talin-vinculin spatial proximities in livers in response to spotted fever group rickettsial and Ebola virus infections.Lab Invest. 2020 Aug;100(8):1030-1041. doi: 10.1038/s41374-020-0420-9. Epub 2020 Apr 1. Lab Invest. 2020. PMID: 32238906 Free PMC article.
-
Spatial Chemoproteomics for Mapping the Active Proteome.Isr J Chem. 2023 Mar;63(3-4):e202200104. doi: 10.1002/ijch.202200104. Epub 2023 Jan 4. Isr J Chem. 2023. PMID: 38046285 Free PMC article.
-
Chemical probes for spatially resolved measurement of active enzymes in single cells.Methods Enzymol. 2019;628:243-262. doi: 10.1016/bs.mie.2019.06.017. Epub 2019 Jul 12. Methods Enzymol. 2019. PMID: 31668232 Free PMC article.
-
A Concise, Modular Antibody-Oligonucleotide Conjugation Strategy Based on Disuccinimidyl Ester Activation Chemistry.Chembiochem. 2019 Jun 14;20(12):1599-1605. doi: 10.1002/cbic.201900027. Epub 2019 May 3. Chembiochem. 2019. PMID: 30767357 Free PMC article.
-
Non-specific interactions of antibody-oligonucleotide conjugates with living cells.Sci Rep. 2021 Mar 15;11(1):5881. doi: 10.1038/s41598-021-85352-w. Sci Rep. 2021. PMID: 33723336 Free PMC article.
References
-
- Pawson T, Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev. 2000;14:1027–1047. - PubMed
-
- Walsh C. Posttranslational Modification of Proteins: Expanding Nature’s Inventory. Englewood: Roberts and Co. Publishers; 2006.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

