CREBBP and p300 lysine acetyl transferases in the DNA damage response
- PMID: 29170789
- PMCID: PMC11105205
- DOI: 10.1007/s00018-017-2717-4
CREBBP and p300 lysine acetyl transferases in the DNA damage response
Abstract
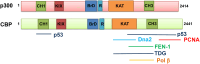
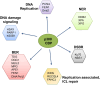
The CREB-binding protein (CREBBP, or in short CBP) and p300 are lysine (K) acetyl transferases (KAT) belonging to the KAT3 family of proteins known to modify histones, as well as non-histone proteins, thereby regulating chromatin accessibility and transcription. Previous studies have indicated a tumor suppressor function for these enzymes. Recently, they have been found to acetylate key factors involved in DNA replication, and in different DNA repair processes, such as base excision repair, nucleotide excision repair, and non-homologous end joining. The growing list of CBP/p300 substrates now includes factors involved in DNA damage signaling, and in other pathways of the DNA damage response (DDR). This review will focus on the role of CBP and p300 in the acetylation of DDR proteins, and will discuss how this post-translational modification influences their functions at different levels, including catalytic activity, DNA binding, nuclear localization, and protein turnover. In addition, we will exemplify how these functions may be necessary to efficiently coordinate the spatio-temporal response to DNA damage. CBP and p300 may contribute to genome stability by fine-tuning the functions of DNA damage signaling and DNA repair factors, thereby expanding their role as tumor suppressors.
Keywords: DNA repair; DNA repair enzymes; DNA replication; Post-translational modification; Protein acetylation.
Figures
Similar articles
-
CBP and p300 acetylate PCNA to link its degradation with nucleotide excision repair synthesis.Nucleic Acids Res. 2014 Jul;42(13):8433-48. doi: 10.1093/nar/gku533. Epub 2014 Jun 17. Nucleic Acids Res. 2014. PMID: 24939902 Free PMC article.
-
p300/CBP acetyl transferases interact with and acetylate the nucleotide excision repair factor XPG.DNA Repair (Amst). 2012 Oct 1;11(10):844-52. doi: 10.1016/j.dnarep.2012.08.001. Epub 2012 Sep 3. DNA Repair (Amst). 2012. PMID: 22954786
-
Mutations in CREBBP and EP300 genes affect DNA repair of oxidative damage in Rubinstein-Taybi syndrome cells.Carcinogenesis. 2020 May 14;41(3):257-266. doi: 10.1093/carcin/bgz149. Carcinogenesis. 2020. PMID: 31504229
-
Endocrine disrupting chemicals may deregulate DNA repair through estrogen receptor mediated seizing of CBP/p300 acetylase.J Endocrinol Invest. 2020 Sep;43(9):1189-1196. doi: 10.1007/s40618-020-01241-5. Epub 2020 Apr 6. J Endocrinol Invest. 2020. PMID: 32253726 Review.
-
An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
Cited by
-
Insights into the Role of the Microbiota and of Short-Chain Fatty Acids in Rubinstein-Taybi Syndrome.Int J Mol Sci. 2021 Mar 31;22(7):3621. doi: 10.3390/ijms22073621. Int J Mol Sci. 2021. PMID: 33807238 Free PMC article.
-
Beta human papillomavirus 8E6 promotes alternative end joining.Elife. 2023 Jan 24;12:e81923. doi: 10.7554/eLife.81923. Elife. 2023. PMID: 36692284 Free PMC article.
-
Clinically Actionable Insights into Initial and Matched Recurrent Glioblastomas to Inform Novel Treatment Approaches.J Oncol. 2019 Dec 31;2019:4878547. doi: 10.1155/2019/4878547. eCollection 2019. J Oncol. 2019. PMID: 32082376 Free PMC article.
-
Regulation of alternative splicing by p300-mediated acetylation of splicing factors.RNA. 2019 Jul;25(7):813-824. doi: 10.1261/rna.069856.118. Epub 2019 Apr 15. RNA. 2019. PMID: 30988101 Free PMC article.
-
Discovery of a peptide proteolysis-targeting chimera (PROTAC) drug of p300 for prostate cancer therapy.EBioMedicine. 2024 Jul;105:105212. doi: 10.1016/j.ebiom.2024.105212. Epub 2024 Jul 1. EBioMedicine. 2024. PMID: 38954976 Free PMC article.
References
-
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

