Structural analyses of key features in the KANK1·KIF21A complex yield mechanistic insights into the cross-talk between microtubules and the cell cortex
- PMID: 29158259
- PMCID: PMC5766899
- DOI: 10.1074/jbc.M117.816017
Structural analyses of key features in the KANK1·KIF21A complex yield mechanistic insights into the cross-talk between microtubules and the cell cortex
Abstract
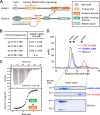
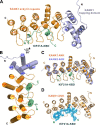
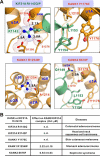
The cross-talk between dynamic microtubules and the cell cortex plays important roles in cell division, polarity, and migration. A critical adaptor that links the plus ends of microtubules with the cell cortex is the KANK N-terminal motif and ankyrin repeat domains 1 (KANK1)/kinesin family member 21A (KIF21A) complex. Genetic defects in these two proteins are associated with various cancers and developmental diseases, such as congenital fibrosis of the extraocular muscles type 1. However, the molecular mechanism governing the KANK1/KIF21A interaction and the role of the conserved ankyrin (ANK) repeats in this interaction are still unclear. In this study, we present the crystal structure of the KANK1·KIF21A complex at 2.1 Å resolution. The structure, together with biochemical studies, revealed that a five-helix-bundle-capping domain immediately preceding the ANK repeats of KANK1 forms a structural and functional supramodule with its ANK repeats in binding to an evolutionarily conserved peptide located in the middle of KIF21A. We also show that several missense mutations present in cancer patients are located at the interface of the KANK1·KIF21A complex and destabilize its formation. In conclusion, our study elucidates the molecular basis underlying the KANK1/KIF21A interaction and also provides possible mechanistic explanations for the diseases caused by mutations in KANK1 and KIF21A.
Keywords: KANK1/KIF21A complex; ankyrin repeat; cell adhesion; kinesin; microtubule; microtubule-cell adhesion cross-talk; scaffold protein; structural biology.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




Similar articles
-
Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.J Biol Chem. 2018 Jan 12;293(2):557-566. doi: 10.1074/jbc.M117.817494. Epub 2017 Nov 28. J Biol Chem. 2018. PMID: 29183992 Free PMC article.
-
Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion.J Biol Chem. 2018 Feb 9;293(6):1944-1956. doi: 10.1074/jbc.M117.815779. Epub 2017 Dec 7. J Biol Chem. 2018. PMID: 29217769 Free PMC article.
-
A major mutation of KIF21A associated with congenital fibrosis of the extraocular muscles type 1 (CFEOM1) enhances translocation of Kank1 to the membrane.Biochem Biophys Res Commun. 2009 Sep 4;386(4):639-44. doi: 10.1016/j.bbrc.2009.06.109. Epub 2009 Jun 24. Biochem Biophys Res Commun. 2009. PMID: 19559006
-
KANK family proteins in cancer.Int J Biochem Cell Biol. 2021 Feb;131:105903. doi: 10.1016/j.biocel.2020.105903. Epub 2020 Dec 10. Int J Biochem Cell Biol. 2021. PMID: 33309958 Review.
-
Kank proteins: structure, functions and diseases.Cell Mol Life Sci. 2009 Aug;66(16):2651-9. doi: 10.1007/s00018-009-0038-y. Epub 2009 May 12. Cell Mol Life Sci. 2009. PMID: 19554261 Free PMC article. Review.
Cited by
-
Kank1 and Ki67 expression are associated with poor prognosis in human pulmonary adenocarcinoma.Int J Clin Exp Pathol. 2020 Sep 1;13(9):2312-2318. eCollection 2020. Int J Clin Exp Pathol. 2020. PMID: 33042336 Free PMC article.
-
Muscleblind-like proteins use modular domains to localize RNAs by riding kinesins and docking to membranes.Nat Commun. 2023 Jun 9;14(1):3427. doi: 10.1038/s41467-023-38923-6. Nat Commun. 2023. PMID: 37296096 Free PMC article.
-
Kinesin-4 KIF21B limits microtubule growth to allow rapid centrosome polarization in T cells.Elife. 2020 Dec 21;9:e62876. doi: 10.7554/eLife.62876. Elife. 2020. PMID: 33346730 Free PMC article.
-
Talin2 and KANK2 functionally interact to regulate microtubule dynamics, paclitaxel sensitivity and cell migration in the MDA-MB-435S melanoma cell line.Cell Mol Biol Lett. 2023 Jul 17;28(1):56. doi: 10.1186/s11658-023-00473-6. Cell Mol Biol Lett. 2023. PMID: 37460977 Free PMC article.
-
Kif21a deficiency leads to impaired glomerular filtration barrier function.Sci Rep. 2023 Nov 6;13(1):19161. doi: 10.1038/s41598-023-46270-1. Sci Rep. 2023. PMID: 37932480 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

