Targeting protein quality control pathways in breast cancer
- PMID: 29145850
- PMCID: PMC5689203
- DOI: 10.1186/s12915-017-0449-4
Targeting protein quality control pathways in breast cancer
Abstract
The efficient production, folding, and secretion of proteins is critical for cancer cell survival. However, cancer cells thrive under stress conditions that damage proteins, so many cancer cells overexpress molecular chaperones that facilitate protein folding and target misfolded proteins for degradation via the ubiquitin-proteasome or autophagy pathway. Stress response pathway induction is also important for cancer cell survival. Indeed, validated targets for anti-cancer treatments include molecular chaperones, components of the unfolded protein response, the ubiquitin-proteasome system, and autophagy. We will focus on links between breast cancer and these processes, as well as the development of drug resistance, relapse, and treatment.
Conflict of interest statement
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
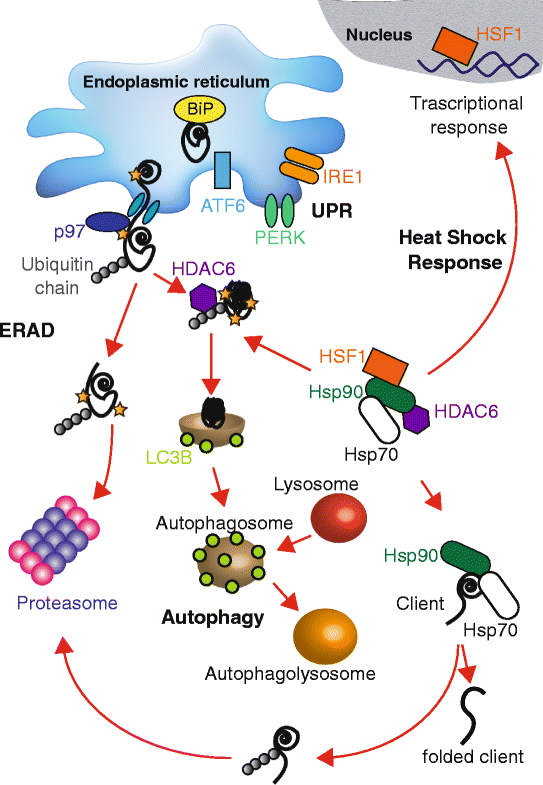
Similar articles
-
Autophagy: links with the proteasome.Curr Opin Cell Biol. 2010 Apr;22(2):192-8. doi: 10.1016/j.ceb.2009.11.002. Epub 2009 Dec 2. Curr Opin Cell Biol. 2010. PMID: 19962293
-
Role of the aggresome pathway in cancer: targeting histone deacetylase 6-dependent protein degradation.Cancer Res. 2008 Apr 15;68(8):2557-60. doi: 10.1158/0008-5472.CAN-07-5989. Cancer Res. 2008. PMID: 18413721 Review.
-
The triage of damaged proteins: degradation by the ubiquitin-proteasome pathway or repair by molecular chaperones.FASEB J. 2006 Apr;20(6):741-3. doi: 10.1096/fj.05-5080fje. Epub 2006 Feb 9. FASEB J. 2006. PMID: 16469848 Free PMC article.
-
Neuronal DnaJ proteins HSJ1a and HSJ1b: a role in linking the Hsp70 chaperone machine to the ubiquitin-proteasome system?Biochem Soc Trans. 2004 Aug;32(Pt 4):640-2. doi: 10.1042/BST0320640. Biochem Soc Trans. 2004. PMID: 15270696
-
Collective roles of molecular chaperones in protein degradation pathways associated with neurodegenerative diseases.Curr Pharm Biotechnol. 2010 Feb;11(2):180-7. doi: 10.2174/138920110790909740. Curr Pharm Biotechnol. 2010. PMID: 20166963 Review.
Cited by
-
Genetic evidence supporting potential causal roles of EIF4 family in breast cancer: a two-sample randomized Mendelian study.Sci Rep. 2024 Aug 30;14(1):20191. doi: 10.1038/s41598-024-71059-1. Sci Rep. 2024. PMID: 39215053 Free PMC article.
-
Unique integrated stress response sensors regulate cancer cell susceptibility when Hsp70 activity is compromised.Elife. 2021 Jun 28;10:e64977. doi: 10.7554/eLife.64977. Elife. 2021. PMID: 34180400 Free PMC article.
-
Compensatory increases of select proteostasis networks after Hsp70 inhibition in cancer cells.J Cell Sci. 2018 Sep 5;131(17):jcs217760. doi: 10.1242/jcs.217760. J Cell Sci. 2018. PMID: 30131440 Free PMC article.
-
Non-Essential Amino Acid Availability Influences Proteostasis and Breast Cancer Cell Survival During Proteotoxic Stress.Mol Cancer Res. 2023 Jul 5;21(7):675-690. doi: 10.1158/1541-7786.MCR-22-0843. Mol Cancer Res. 2023. PMID: 36961392 Free PMC article.
-
Aggresomes predict poor outcomes and implicate proteostasis in the pathogenesis of pediatric choroid plexus tumors.J Neurooncol. 2021 Mar;152(1):67-78. doi: 10.1007/s11060-020-03694-3. Epub 2021 Jan 26. J Neurooncol. 2021. PMID: 33501605 Free PMC article.
References
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. - PubMed
-
- Howell A, Howell SJ, Evans DG. New approaches to the endocrine prevention and treatment of breast cancer. Cancer Chemother Pharmacol. 2003;52(Suppl 1):S39–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

