Respiratory syncytial virus induces NRF2 degradation through a promyelocytic leukemia protein - ring finger protein 4 dependent pathway
- PMID: 29107745
- PMCID: PMC5699968
- DOI: 10.1016/j.freeradbiomed.2017.10.380
Respiratory syncytial virus induces NRF2 degradation through a promyelocytic leukemia protein - ring finger protein 4 dependent pathway
Abstract
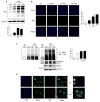
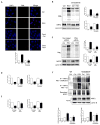
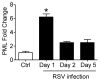
Respiratory syncytial virus (RSV) is the most important cause of viral acute respiratory tract infections and hospitalizations in children, for which no vaccine or specific treatments are available. RSV causes airway mucosa inflammation and cellular oxidative damage by triggering production of reactive oxygen species and by inhibiting at the same time expression of antioxidant enzymes, via degradation of the transcription factor NF-E2-related factor 2 (NRF2). RSV infection induces NRF2 deacetylation, ubiquitination, and degradation through a proteasome-dependent pathway. Although degradation via KEAP1 is the most common mechanism, silencing KEAP1 expression did not rescue NRF2 levels during RSV infection. We found that RSV-induced NRF2 degradation occurs in an SUMO-specific E3 ubiquitin ligase - RING finger protein 4 (RNF4)-dependent manner. NRF2 is progressively SUMOylated in RSV infection and either blocking SUMOylation or silencing RNF4 expression rescued both NRF2 nuclear levels and transcriptional activity. RNF4 associates with promyelocytic leukemia - nuclear bodies (PML-NBs). RSV infection induces the expression of PML and PML-NBs formation in an interferon (INF)-dependent manner and also induces NRF2 - PMN-NBs association. Inhibition of PML-NB formation by blocking IFN pathway or silencing PML expression resulted in a significant reduction of RSV-associated NRF2 degradation and increased antioxidant enzyme expression, identifying the RNF4-PML pathway as a key regulator of antioxidant defenses in the course of viral infection.
Keywords: Antioxidant enzymes; KEAP1; NRF2; PML nuclear bodies; RNF4; Respiratory syncytial virus; SUMOylation; Ubiquitination.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest: none
Figures



Similar articles
-
Trafficking of the transcription factor Nrf2 to promyelocytic leukemia-nuclear bodies: implications for degradation of NRF2 in the nucleus.J Biol Chem. 2013 May 17;288(20):14569-14583. doi: 10.1074/jbc.M112.437392. Epub 2013 Mar 29. J Biol Chem. 2013. PMID: 23543742 Free PMC article.
-
Inhibiting ubiquitination causes an accumulation of SUMOylated newly synthesized nuclear proteins at PML bodies.J Biol Chem. 2019 Oct 18;294(42):15218-15234. doi: 10.1074/jbc.RA119.009147. Epub 2019 Jul 8. J Biol Chem. 2019. PMID: 31285264 Free PMC article.
-
Effect of SUMO-SIM Interaction on the ICP0-Mediated Degradation of PML Isoform II and Its Associated Proteins in Herpes Simplex Virus 1 Infection.J Virol. 2020 Jun 1;94(12):e00470-20. doi: 10.1128/JVI.00470-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32295906 Free PMC article.
-
Targeting the KEAP1-NRF2 System to Prevent Kidney Disease Progression.Am J Nephrol. 2017;45(6):473-483. doi: 10.1159/000475890. Epub 2017 May 13. Am J Nephrol. 2017. PMID: 28502971 Review.
-
A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics.Int J Biol Sci. 2010 Jan 12;6(1):51-67. doi: 10.7150/ijbs.6.51. Int J Biol Sci. 2010. PMID: 20087442 Free PMC article. Review.
Cited by
-
Hydrogen Sulfide Donor GYY4137 Rescues NRF2 Activation in Respiratory Syncytial Virus Infection.Antioxidants (Basel). 2022 Jul 21;11(7):1410. doi: 10.3390/antiox11071410. Antioxidants (Basel). 2022. PMID: 35883901 Free PMC article.
-
Endothelial Dysfunction through Oxidatively Generated Epigenetic Mark in Respiratory Viral Infections.Cells. 2021 Nov 7;10(11):3067. doi: 10.3390/cells10113067. Cells. 2021. PMID: 34831290 Free PMC article. Review.
-
Modeling Human Respiratory Syncytial Virus (RSV) Infection: Recent Contributions and Future Directions Using the Calf Model of Bovine RSV Disease.J Immunol. 2023 Oct 15;211(8):1180-1186. doi: 10.4049/jimmunol.2300260. J Immunol. 2023. PMID: 37782855 Free PMC article. Review.
-
Anti-Viral Potential and Modulation of Nrf2 by Curcumin: Pharmacological Implications.Antioxidants (Basel). 2020 Dec 4;9(12):1228. doi: 10.3390/antiox9121228. Antioxidants (Basel). 2020. PMID: 33291560 Free PMC article. Review.
-
Bioactivity, bioavailability, and gut microbiota transformations of dietary phenolic compounds: implications for COVID-19.J Nutr Biochem. 2021 Nov;97:108787. doi: 10.1016/j.jnutbio.2021.108787. Epub 2021 Jun 2. J Nutr Biochem. 2021. PMID: 34089819 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

