TumorFusions: an integrative resource for cancer-associated transcript fusions
- PMID: 29099951
- PMCID: PMC5753333
- DOI: 10.1093/nar/gkx1018
TumorFusions: an integrative resource for cancer-associated transcript fusions
Abstract
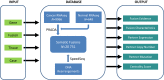
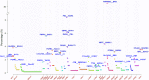
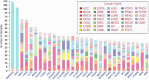
Gene fusion represents a class of molecular aberrations in cancer and has been exploited for therapeutic purposes. In this paper we describe TumorFusions, a data portal that catalogues 20 731 gene fusions detected in 9966 well characterized cancer samples and 648 normal specimens from The Cancer Genome Atlas (TCGA). The portal spans 33 cancer types in TCGA. Fusion transcripts were identified via a uniform pipeline, including filtering against a list of 3838 transcript fusions detected in a panel of 648 non-neoplastic samples. Fusions were mapped to somatic DNA rearrangements identified using whole genome sequencing data from 561 cancer samples as a means of validation. We observed that 65% of transcript fusions were associated with a chromosomal alteration, which is annotated in the portal. Other features of the portal include links to SNP array-based copy number levels and mutational patterns, exon and transcript level expressions of the partner genes, and a network-based centrality score for prioritizing functional fusions. Our portal aims to be a broadly applicable and user friendly resource for cancer gene annotation and is publicly available at http://www.tumorfusions.org.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
FusionGDB: fusion gene annotation DataBase.Nucleic Acids Res. 2019 Jan 8;47(D1):D994-D1004. doi: 10.1093/nar/gky1067. Nucleic Acids Res. 2019. PMID: 30407583 Free PMC article.
-
The landscape and therapeutic relevance of cancer-associated transcript fusions.Oncogene. 2015 Sep 10;34(37):4845-54. doi: 10.1038/onc.2014.406. Epub 2014 Dec 15. Oncogene. 2015. PMID: 25500544 Free PMC article.
-
annoFuse: an R Package to annotate, prioritize, and interactively explore putative oncogenic RNA fusions.BMC Bioinformatics. 2020 Dec 14;21(1):577. doi: 10.1186/s12859-020-03922-7. BMC Bioinformatics. 2020. PMID: 33317447 Free PMC article.
-
Chromosomal aberrations in solid tumors.Prog Mol Biol Transl Sci. 2010;95:55-94. doi: 10.1016/B978-0-12-385071-3.00004-6. Prog Mol Biol Transl Sci. 2010. PMID: 21075329 Review.
-
Gene fusions in tumourigenesis with particular reference to ovarian cancer.J Med Genet. 2021 Dec;58(12):789-795. doi: 10.1136/jmedgenet-2021-108010. Epub 2021 Aug 30. J Med Genet. 2021. PMID: 34462289 Review.
Cited by
-
Novornabreak: Local Assembly for Novel Splice Junction and Fusion Transcript Detection from RNA-Seq Data.J Bioinform Syst Biol. 2023;6(2):74-81. doi: 10.26502/jbsb.5107050. Epub 2023 Apr 4. J Bioinform Syst Biol. 2023. PMID: 39301431 Free PMC article.
-
ESR1 Fusions Invoke Breast Cancer Subtype-Dependent Enrichment of Ligand-Independent Oncogenic Signatures and Phenotypes.Endocrinology. 2024 Aug 27;165(10):bqae111. doi: 10.1210/endocr/bqae111. Endocrinology. 2024. PMID: 39207954 Free PMC article.
-
Comprehensive molecular characterization of TFE3-rearranged renal cell carcinoma.Exp Mol Med. 2024 Aug;56(8):1807-1815. doi: 10.1038/s12276-024-01291-2. Epub 2024 Aug 1. Exp Mol Med. 2024. PMID: 39085357 Free PMC article.
-
WNT inhibitory factor 1 (WIF1) is a novel fusion partner of RUNX family transcription factor 1 (RUNX1) in acute myeloid leukemia with t(12;21)(q14;q22).J Hematop. 2024 Jul 27. doi: 10.1007/s12308-024-00597-4. Online ahead of print. J Hematop. 2024. PMID: 39066949
-
Identifying new cancer genes based on the integration of annotated gene sets via hypergraph neural networks.Bioinformatics. 2024 Jun 28;40(Suppl 1):i511-i520. doi: 10.1093/bioinformatics/btae257. Bioinformatics. 2024. PMID: 38940121 Free PMC article.
References
-
- Mertens F., Johansson B., Fioretos T., Mitelman F.. The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer. 2015; 15:371–381. - PubMed
-
- Kloosterman W.P., Coebergh van den Braak R.R.J., Pieterse M., van Roosmalen M.J., Sieuwerts A.M., Stangl C., Brunekreef R., Lalmahomed Z.S., Ooft S., van Galen A. et al. . A systematic analysis of oncogenic gene fusions in primary colon cancer. Cancer Res. 2017; 77:3814–3822. - PubMed
-
- Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R. et al. . Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005; 310:644–648. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

