Endosomal phosphatidylserine is critical for the YAP signalling pathway in proliferating cells
- PMID: 29093443
- PMCID: PMC5665887
- DOI: 10.1038/s41467-017-01255-3
Endosomal phosphatidylserine is critical for the YAP signalling pathway in proliferating cells
Abstract
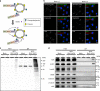
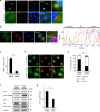
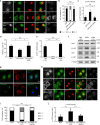
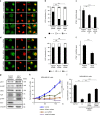
Yes-associated protein (YAP) is a recently discovered growth-promoting transcription coactivator that has been shown to regulate the malignancy of various cancers. How YAP is regulated is not fully understood. Here, we show that one of the factors regulating YAP is phosphatidylserine (PS) in recycling endosomes (REs). We use proximity biotinylation to find proteins proximal to PS. Among these proteins are YAP and multiple proteins related to YAP signalling. Knockdown of ATP8A1 (an RE PS-flippase) or evectin-2 (an RE-resident protein) and masking of PS in the cytoplasmic leaflet of membranes, all suppress nuclear localization of YAP and YAP-dependent transcription. ATP8A1 knockdown increases the phosphorylated (activated) form of Lats1 that phosphorylates and inactivates YAP, whereas evectin-2 knockdown reduces the ubiquitination and increased the level of Lats1. The proliferation of YAP-dependent metastatic cancer cells is suppressed by knockdown of ATP8A1 or evectin-2. These results suggest a link between a membrane phospholipid and cell proliferation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Similar articles
-
AP-3-dependent targeting of flippase ATP8A1 to lamellar bodies suppresses activation of YAP in alveolar epithelial type 2 cells.Proc Natl Acad Sci U S A. 2021 May 18;118(20):e2025208118. doi: 10.1073/pnas.2025208118. Proc Natl Acad Sci U S A. 2021. PMID: 33990468 Free PMC article.
-
PPP1R12A is a recycling endosomal phosphatase that facilitates YAP activation.Sci Rep. 2023 Nov 13;13(1):19740. doi: 10.1038/s41598-023-47138-0. Sci Rep. 2023. PMID: 37957190 Free PMC article.
-
Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase.EMBO J. 2015 Mar 4;34(5):669-88. doi: 10.15252/embj.201489703. Epub 2015 Jan 16. EMBO J. 2015. PMID: 25595798 Free PMC article.
-
Furry protein suppresses nuclear localization of yes-associated protein (YAP) by activating NDR kinase and binding to YAP.J Biol Chem. 2020 Mar 6;295(10):3017-3028. doi: 10.1074/jbc.RA119.010783. Epub 2020 Jan 29. J Biol Chem. 2020. PMID: 31996378 Free PMC article.
-
A Role of Phosphatidylserine in the Function of Recycling Endosomes.Front Cell Dev Biol. 2021 Dec 24;9:783857. doi: 10.3389/fcell.2021.783857. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 35004683 Free PMC article. Review.
Cited by
-
SREBP1 silencing inhibits the proliferation and motility of human esophageal squamous carcinoma cells via the Wnt/β-catenin signaling pathway.Oncol Lett. 2020 Sep;20(3):2855-2869. doi: 10.3892/ol.2020.11853. Epub 2020 Jul 10. Oncol Lett. 2020. PMID: 32765792 Free PMC article.
-
AP-3-dependent targeting of flippase ATP8A1 to lamellar bodies suppresses activation of YAP in alveolar epithelial type 2 cells.Proc Natl Acad Sci U S A. 2021 May 18;118(20):e2025208118. doi: 10.1073/pnas.2025208118. Proc Natl Acad Sci U S A. 2021. PMID: 33990468 Free PMC article.
-
Distribution, dynamics and functional roles of phosphatidylserine within the cell.Cell Commun Signal. 2019 Oct 15;17(1):126. doi: 10.1186/s12964-019-0438-z. Cell Commun Signal. 2019. PMID: 31615534 Free PMC article. Review.
-
TurboID-EV: Proteomic Mapping of Recipient Cellular Proteins Proximal to Small Extracellular Vesicles.Anal Chem. 2023 Sep 26;95(38):14159-14164. doi: 10.1021/acs.analchem.3c01015. Epub 2023 Sep 14. Anal Chem. 2023. PMID: 37709279 Free PMC article.
-
Pathogenesis and Therapy of Hermansky-Pudlak Syndrome (HPS)-Associated Pulmonary Fibrosis.Int J Mol Sci. 2024 Oct 19;25(20):11270. doi: 10.3390/ijms252011270. Int J Mol Sci. 2024. PMID: 39457053 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

