Hijacking of the Ubiquitin/Proteasome Pathway by the HIV Auxiliary Proteins
- PMID: 29088112
- PMCID: PMC5707529
- DOI: 10.3390/v9110322
Hijacking of the Ubiquitin/Proteasome Pathway by the HIV Auxiliary Proteins
Abstract
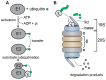
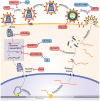
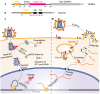
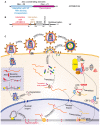
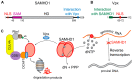
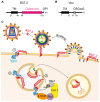
The ubiquitin-proteasome system (UPS) ensures regulation of the protein pool in the cell by ubiquitination of proteins followed by their degradation by the proteasome. It plays a central role in the cell under normal physiological conditions as well as during viral infections. On the one hand, the UPS can be used by the cell to degrade viral proteins, thereby restricting the viral infection. On the other hand, it can also be subverted by the virus to its own advantage, notably to induce degradation of cellular restriction factors. This makes the UPS a central player in viral restriction and counter-restriction. In this respect, the human immunodeficiency viruses (HIV-1 and 2) represent excellent examples. Indeed, many steps of the HIV life cycle are restricted by cellular proteins, some of which are themselves components of the UPS. However, HIV itself hijacks the UPS to mediate defense against several cellular restriction factors. For example, the HIV auxiliary proteins Vif, Vpx and Vpu counteract specific restriction factors by the recruitment of cellular UPS components. In this review, we describe the interplay between HIV and the UPS to illustrate its role in the restriction of viral infections and its hijacking by viral proteins for counter-restriction.
Keywords: APOBEC; BST2/Tetherin; HIV; March8; SAMHD1; TRIM5α; proteasome; restriction factors; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Structural basis of lentiviral subversion of a cellular protein degradation pathway.Nature. 2014 Jan 9;505(7482):234-8. doi: 10.1038/nature12815. Epub 2013 Dec 15. Nature. 2014. PMID: 24336198 Free PMC article.
-
Role of the endosomal ESCRT machinery in HIV-1 Vpu-induced down-regulation of BST2/tetherin.Curr HIV Res. 2012 Jun;10(4):315-20. doi: 10.2174/157016212800792414. Curr HIV Res. 2012. PMID: 22524180 Review.
-
Characterization of E3 ligases involved in lysosomal sorting of the HIV-1 restriction factor BST2.J Cell Sci. 2017 May 1;130(9):1596-1611. doi: 10.1242/jcs.195412. Epub 2017 Mar 20. J Cell Sci. 2017. PMID: 28320822 Free PMC article.
-
Binding to DCAF1 distinguishes TASOR and SAMHD1 degradation by HIV-2 Vpx.PLoS Pathog. 2021 Oct 26;17(10):e1009609. doi: 10.1371/journal.ppat.1009609. eCollection 2021 Oct. PLoS Pathog. 2021. PMID: 34699574 Free PMC article.
-
Role of the Ubiquitin Proteasome System (UPS) in the HIV-1 Life Cycle.Int J Mol Sci. 2019 Jun 19;20(12):2984. doi: 10.3390/ijms20122984. Int J Mol Sci. 2019. PMID: 31248071 Free PMC article. Review.
Cited by
-
Help or Hinder: Protein Host Factors That Impact HIV-1 Replication.Viruses. 2024 Aug 10;16(8):1281. doi: 10.3390/v16081281. Viruses. 2024. PMID: 39205255 Free PMC article. Review.
-
Proteasomal Degradation Machinery: Favorite Target of HIV-1 Proteins.Front Microbiol. 2018 Nov 21;9:2738. doi: 10.3389/fmicb.2018.02738. eCollection 2018. Front Microbiol. 2018. PMID: 30524389 Free PMC article. Review.
-
Regulation of Viral Restriction by Post-Translational Modifications.Viruses. 2021 Nov 1;13(11):2197. doi: 10.3390/v13112197. Viruses. 2021. PMID: 34835003 Free PMC article. Review.
-
Ubiquitin E3 Ligase c-Cbl Is a Host Negative Regulator of Nef Protein of HIV-1.Front Microbiol. 2020 Nov 19;11:597972. doi: 10.3389/fmicb.2020.597972. eCollection 2020. Front Microbiol. 2020. PMID: 33329486 Free PMC article.
-
Molecular basis of cullin-3 (Cul3) ubiquitin ligase subversion by vaccinia virus protein A55.J Biol Chem. 2019 Apr 19;294(16):6416-6429. doi: 10.1074/jbc.RA118.006561. Epub 2019 Feb 28. J Biol Chem. 2019. PMID: 30819806 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

