A neuroprotective astrocyte state is induced by neuronal signal EphB1 but fails in ALS models
- PMID: 29079839
- PMCID: PMC5660125
- DOI: 10.1038/s41467-017-01283-z
A neuroprotective astrocyte state is induced by neuronal signal EphB1 but fails in ALS models
Abstract
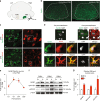
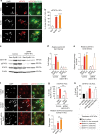
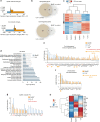
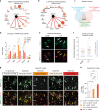
Astrocyte responses to neuronal injury may be beneficial or detrimental to neuronal recovery, but the mechanisms that determine these different responses are poorly understood. Here we show that ephrin type-B receptor 1 (EphB1) is upregulated in injured motor neurons, which in turn can activate astrocytes through ephrin-B1-mediated stimulation of signal transducer and activator of transcription-3 (STAT3). Transcriptional analysis shows that EphB1 induces a protective and anti-inflammatory signature in astrocytes, partially linked to the STAT3 network. This is distinct from the response evoked by interleukin (IL)-6 that is known to induce both pro inflammatory and anti-inflammatory processes. Finally, we demonstrate that the EphB1-ephrin-B1 pathway is disrupted in human stem cell derived astrocyte and mouse models of amyotrophic lateral sclerosis (ALS). Our work identifies an early neuronal help-me signal that activates a neuroprotective astrocytic response, which fails in ALS, and therefore represents an attractive therapeutic target.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Angiogenin induces modifications in the astrocyte secretome: relevance to amyotrophic lateral sclerosis.J Proteomics. 2013 Oct 8;91:274-85. doi: 10.1016/j.jprot.2013.07.028. Epub 2013 Aug 3. J Proteomics. 2013. PMID: 23920243
-
Activation of STAT3 and inhibitory effects of pioglitazone on STAT3 activity in a mouse model of SOD1-mutated amyotrophic lateral sclerosis.Neuropathology. 2010 Aug;30(4):353-60. doi: 10.1111/j.1440-1789.2009.01078.x. Epub 2009 Nov 18. Neuropathology. 2010. PMID: 19925559
-
Reactive Astrocytes Promote ALS-like Degeneration and Intracellular Protein Aggregation in Human Motor Neurons by Disrupting Autophagy through TGF-β1.Stem Cell Reports. 2017 Aug 8;9(2):667-680. doi: 10.1016/j.stemcr.2017.06.008. Epub 2017 Jul 14. Stem Cell Reports. 2017. PMID: 28712846 Free PMC article.
-
Complexity of astrocyte-motor neuron interactions in amyotrophic lateral sclerosis.Neurodegener Dis. 2005;2(3-4):139-46. doi: 10.1159/000089619. Neurodegener Dis. 2005. PMID: 16909019 Review.
-
The multi-dimensional roles of astrocytes in ALS.Neurosci Res. 2018 Jan;126:31-38. doi: 10.1016/j.neures.2017.09.011. Epub 2017 Oct 17. Neurosci Res. 2018. PMID: 29054467 Review.
Cited by
-
Decoding the relationship between ageing and amyotrophic lateral sclerosis: a cellular perspective.Brain. 2020 Apr 1;143(4):1057-1072. doi: 10.1093/brain/awz360. Brain. 2020. PMID: 31851317 Free PMC article. Review.
-
Delineating Astrocytic Cytokine Responses in a Human Stem Cell Model of Neural Trauma.J Neurotrauma. 2020 Jan 1;37(1):93-105. doi: 10.1089/neu.2019.6480. Epub 2019 Sep 18. J Neurotrauma. 2020. PMID: 31452443 Free PMC article.
-
Astrocytes: Dissecting Their Diverse Roles in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia.Cells. 2023 May 23;12(11):1450. doi: 10.3390/cells12111450. Cells. 2023. PMID: 37296571 Free PMC article. Review.
-
Aberrant activation of hippocampal astrocytes causes neuroinflammation and cognitive decline in mice.PLoS Biol. 2024 Jul 11;22(7):e3002687. doi: 10.1371/journal.pbio.3002687. eCollection 2024 Jul. PLoS Biol. 2024. PMID: 38991663 Free PMC article.
-
Glial Cells-The Strategic Targets in Amyotrophic Lateral Sclerosis Treatment.J Clin Med. 2020 Jan 18;9(1):261. doi: 10.3390/jcm9010261. J Clin Med. 2020. PMID: 31963681 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

