TORC1 and TORC2 converge to regulate the SAGA co-activator in response to nutrient availability
- PMID: 29079657
- PMCID: PMC5709762
- DOI: 10.15252/embr.201744942
TORC1 and TORC2 converge to regulate the SAGA co-activator in response to nutrient availability
Abstract
Gene expression regulation is essential for cells to adapt to changes in their environment. Co-activator complexes have well-established roles in transcriptional regulation, but less is known about how they sense and respond to signaling cues. We have previously shown that, in fission yeast, one such co-activator, the SAGA complex, controls gene expression and the switch from proliferation to differentiation in response to nutrient availability. Here, using a combination of genetic, biochemical, and proteomic approaches, we show that SAGA responds to nutrients through the differential phosphorylation of its Taf12 component, downstream of both the TORC1 and TORC2 pathways. Taf12 phosphorylation increases early upon starvation and is controlled by the opposing activities of the PP2A phosphatase, which is activated by TORC1, and the TORC2-activated Gad8AKT kinase. Mutational analyses suggest that Taf12 phosphorylation prevents cells from committing to differentiation until starvation reaches a critical level. Overall, our work reveals that SAGA is a direct target of nutrient-sensing pathways and has uncovered a mechanism by which TORC1 and TORC2 converge to control gene expression and cell fate decisions.
Keywords: SAGA; TOR; differentiation; fission yeast; signal transduction; transcription.
© 2017 The Authors.
Figures
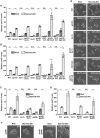
- A–D
Expression of ste11 + (A, C) and mei2 + (B, D) using quantitative RT–PCR of RNA extracted from cells grown either in nutrient rich medium (dark gray) or shifted for 4 h to starvation medium (light gray). Cells of the following genotypes were analyzed: wild‐type isogenic controls (WT), gcn5Δ, tsc1Δ, gcn5Δ tsc1Δ, tsc2Δ, gcn5Δ tsc2Δ, rhb1‐DA4—a constitutively active (CA) rhb1 mutant 34, gcn5Δ rhb1‐DA4, tor2‐L1310P—a CA tor2 mutant 33, gcn5Δ tor2‐L1310P, tor1Δ, gcn5Δ tor1Δ, gad8Δ, and gcn5Δ gad8Δ. act1 + served as a control for normalization across samples. Values from a WT strain grown in rich medium were set at 1 to allow comparisons across culture conditions and mutant strains. Each column represents the mean value of 4 (A, B) or 3 (C, D) independent experiments, overlaid with individual data points and error bars showing the standard error of the mean (SEM). Statistical significance was determined by two‐way ANOVA followed by Bonferroni's multiple comparison tests (n = 4 for A, B; n = 3 for C, D); *P ≤ 0.01.
- E, F
Cells were grown to mid‐log phase either in rich medium or shifted for 8 h to starvation medium. Zygotes and tetrads, which correspond to differentiated cells, were counted under a light microscope. Cells of the following genotypes were analyzed: WT isogenic controls, gcn5Δ, tsc1Δ, gcn5Δ tsc1Δ, rhb1‐DA4, gcn5Δ rhb1‐DA4, gad8Δ, and gcn5Δ gad8Δ. Each value represents the mean percentage and SEM of differentiating cells to the total number of cells, averaged from four independent experiments. At least 200 cells from the indicated genotypes were counted in each experiment. White arrowheads indicate zygotes. Scale bar, 10 μm.

- A, B
4–20% gradient SDS–polyacrylamide gel electrophoresis analysis of SAGA purified from cells grown either in rich medium (R) or starved for 45 min (S). SAGA was purified using endogenously TAP‐tagged Ada1. A fraction of each eluate was loaded and either stained with silver, to detect all proteins (A), or with Pro‐Q® Diamond, which stains phosphorylated proteins (B). A strain without any TAP tag is shown as a negative control for the purification. The graph to the left of the gel in (B) shows the fluorescence intensity of the phospho‐specific stain, which was quantified along the left lane in blue (rich) and the middle lane in red (starved), using ImageJ. The area of one peak, which corresponds to the bands marked with arrowheads and was coined p55, is 1.8‐fold larger in SAGA purified from starved cells, as compared to rich conditions. Below is an anti‐HA immunoblot (IB) of each eluate, to reveal the amount of Ada1‐HA bait recovered. Shown are gels that are representative of three independent experiments. A.U., arbitrary units.

- A
Overview of the quantitative proteomic approaches used to identify differentially phosphorylated peptides, either in crude extracts or in SAGA purifications. Cells were metabolically labeled using a SILAC procedure and grown either in rich medium or shifted to nutrient starvation conditions. Several independent experiments were carried out with forward and reverse labeling schemes (see Materials and Methods for details).
- B
Schematic view of the S. pombe (Sp) Taf12 protein sequence, including, in red, the three differentially phosphorylated Thr, at positions 218, 221, and 283 and, in gray, the histone‐fold domain. Shown below are the starved‐to‐rich SILAC ratios of the signal intensities observed for the Thr218‐Thr221 peptide (Appendix Fig S3) and for the Thr283 peptide (Appendix Fig S4).
- C
Summary of the different taf12 + point mutants that were constructed and analyzed.
- D
FLAG‐tagged Taf12 was purified from cells grown in rich medium (R) or shifted for 45 min to starvation medium (S). Anti‐FLAG immuno‐precipitations (IP) were incubated with and without λ‐phosphatase or its inhibitor and immunoblotted (IB), together with 1% of whole cell extracts (WCE), using an anti‐FLAG antibody.
- E
Phospho‐Taf12 (P‐Taf12), total Taf12, and tubulin levels were quantified from cells grown in rich medium (R) or starved for 45 min (S). P‐Taf12 levels were normalized to those of total Taf12, while total Taf12 levels were normalized to those of tubulin. Data points were individually plotted on the graph and overlaid with the mean and SEM. Signal intensities were quantified from IBs of seven independent experiments. Statistical significance was determined using a t‐test (n = 7, unpaired, two‐tailed); *P ≤ 0.01.
- F
P‐Taf12 and total Taf12 were followed in WT, taf12‐T283A and taf12‐5A mutants, grown in rich medium (R) or starved for 45 min (S).
- G, H
SAGA and TFIID were tandem affinity‐purified using endogenously TAP‐tagged Spt7 (G) or Taf4 (H), from strains containing FLAG‐tagged Taf12, grown in rich medium or starved for 45 min. TAP‐tagged Spt7 and Taf4 were eluted using the TEV protease, releasing a shorter form of each bait (Spt7‐CBP or Taf4‐CBP). Eluates were loaded and immunoblotted (IB) using anti‐FLAG or anti‐CBP antibodies, together with 1% of either whole cell extracts (WCE) or IgG‐Sepharose flow‐through (FT). Shown are IBs that are representative of two independent experiments.
- I
P‐Taf12, total Taf12, and Ser546‐phosphorylated Gad8AKT were followed in WT cells, grown in rich conditions or over a time‐course after a shift to starvation medium. Gad8AKT phosphorylation at Ser546 is a proxy of TORC2 activity in S. pombe.
- J
P‐Taf12 and total Taf12 were followed in WT cells, grown in rich conditions or shifted to different starvation media for 30 min. These include both the removal of the nitrogen source, ammonium chloride (NH4Cl), and the reduction in the carbon source, glucose, from a concentration of 2–0.5%. Alternatively, cells were either only deprived of NH4Cl or only exposed to reduced glucose levels (2–0.5%).
- K
P‐Taf12 and total Taf12 were followed in WT cells, grown in rich conditions or deprived of NH4Cl for 1 h. Then, NH4Cl was added back to the medium for 30 or 60 min.
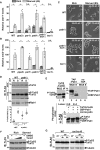
- A, B
Expression of ste11 + (A) and mei2 + (B) using quantitative RT–PCR of RNA extracted from cells grown either in rich medium (dark gray) or starved for 4 h (light gray). Cells of the following genotypes were analyzed: wild‐type isogenic controls (WT), ppa2Δ, par1Δ, pab1Δ, tsc1Δ, and pab1Δ tsc1Δ. act1 + served as a control for normalization across samples. Values from a WT strain grown in rich medium were set at 1 to allow comparisons across culture conditions and mutant strains. Each column represents the mean value of four independent experiments, overlaid with individual data points and SEM. Statistical significance was determined by two‐way ANOVA followed by Bonferroni's multiple comparison tests (n = 4); *P ≤ 0.01.
- C
Cells were grown to mid‐log phase either in rich medium or starved for 8 h. Zygotes and tetrads, which correspond to differentiated cells, were counted under a light microscope. Each value represents the mean percentage and SEM of differentiating cells to the total number of cells, averaged from four independent experiments. At least 200 cells from the indicated genotypes were counted in each experiment. White arrowheads indicate zygotes. Scale bar, 10 μm.
- D
P‐Taf12 was followed by anti‐FLAG IB of protein extracts from WT and pab1Δ strains, grown in rich conditions (R) or starved for 45 min (S). An anti‐Rpb1 IB is shown as a control for loading. The signal intensities of P‐Taf12 and total Taf12 were quantified in each strain and condition. Ratios of P‐Taf12 to total Taf12 were calculated from six independent experiments and individually plotted in a graph below the IBs, together with the mean and SEM. Averaged values from all WT controls grown in rich medium were set at 1 to allow comparisons across culture conditions and strains. Statistical significance was determined by two‐way ANOVA followed by Bonferroni's multiple comparison tests (n = 6; *P < 0.01; # P < 0.05). A short exposure and a long exposure of the FLAG IB are shown to detect total Taf12 and P‐Taf12, respectively, within the linear range of the chemiluminescence signal.
- E
FLAG‐tagged Taf12 and TAP‐tagged Pab1 were affinity‐purified separately from cells grown in rich conditions (upper panels). TAP‐tagged Pab1 was cleaved off from beads using the tobacco etch virus (TEV) protease, releasing a shorter form of Pab1 (CBP‐Pab1) in the eluate (E). CBP‐Pab1 was then mixed with beads containing FLAG‐Taf12 IPs and incubated in a phosphatase buffer, with and without 0.5 μM microcystin. Each reaction was then analyzed by IB and simultaneously probed with anti‐FLAG or anti‐CBP antibodies (lower panel).
- F
Exponentially growing cells were treated for 1 h with Torin‐1, which was added at increasing concentrations, 7 and 21 μM, to rich medium. Dimethylsulfoxide (DMSO) was used as the vehicle and added as a negative control. Anti‐tubulin IBs are shown as a control for loading between samples.
- G
P‐Taf12 was followed by anti‐FLAG IB of protein extracts from WT and tor2‐ts10 strains, grown in rich conditions at 25°C or shifted to 30°C for 4 and 6 h. Anti‐tubulin IBs are shown as a control for loading between samples.
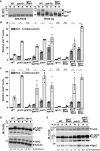
- A
Igo1 phosphorylation (P‐Igo1) was followed by anti‐MYC IB of protein extracts from WT, ppk18Δ, and igo1‐S64A strains, grown in rich conditions (R) or starved for 45 min (S). Igo1 migration was analyzed by electrophoresis of both a 12% SDS–polyacrylamide gel containing the Phos‐tag™ molecule (right panel) and a standard 10% SDS–polyacrylamide gel (left panel). Arrowheads indicate the various phosphorylated isoforms of Igo1‐MYC in each strain and condition. Shown is an IB representative of two independent experiments.
- B, C
Expression of ste11 + (C) and mei2 + (D) using quantitative RT–PCR of RNA extracted from cells grown either in rich medium (dark gray) or starved for 4 h (light gray). Cells of the following genotypes were analyzed: wild‐type isogenic controls (WT), gcn5Δ, ppk18Δ, gcn5Δ ppk18Δ, pab1Δ, pab1Δ ppk18Δ, igo1Δ, igo1‐S64A, gcn5Δ igo1‐S64A, and pab1Δ igo1‐S64A. act1 + served as a control for normalization across samples. Values from a WT strain grown in rich medium were set at 1 to allow comparisons across culture conditions and mutant strains. Each column represents the mean value of four independent experiments, overlaid with individual data points and SEM. Statistical significance was determined by two‐way ANOVA followed by Bonferroni's multiple comparison tests (n = 4); *P ≤ 0.01.
- D
P‐Taf12 was followed by anti‐FLAG IB of protein extracts from WT, ppk18Δ, and igo1Δ strains, grown in rich conditions (R) or starved for 45 min (S). An anti‐tubulin IB is shown as a control for loading.
- E
P‐Taf12 was followed in WT, igoS64A, pab1Δ, and pab1Δ igoS64A strains, grown in rich conditions (R) or starved for 45 min (S). An anti‐Rpb1 IB is shown as a control for loading.

P‐Taf12 was followed by anti‐FLAG IB of protein extracts from WT, tor1Δ, and gad8Δ strains, grown in rich conditions (R) or starved for 45 min (S). An anti‐tubulin IB is shown as a control for loading. The signal intensities of P‐Taf12 and total Taf12 were quantified in each strain and condition. P‐Taf12 to Taf2 ratios were calculated from three independent experiments and individually plotted in a graph below the IBs, together with the mean and SEM. Averaged values from all WT controls grown in rich medium were set at 1 to allow comparisons across culture conditions and mutant strains. Statistical significance was determined by two‐way ANOVA followed by Bonferroni's multiple comparison tests (n = 3); *P ≤ 0.01.
P‐Taf12 was followed by anti‐FLAG IB of protein extracts from sty1Δ cells (left panel) or ssp2Δ cells (right panel), grown in rich conditions (R) or starved for 45 min (S). An anti‐tubulin IB is shown as a control for loading. Shown are IBs that are representative of two independent experiments.
TAP‐tagged Gad8 was immuno‐precipitated using IgG‐Sepharose beads (IP) from strains containing FLAG‐tagged Taf12, grown either in rich medium (left panels) or starved for 45 min (right panels). TAP‐tagged Gad8 was eluted from beads using the TEV protease, releasing a shorter form of Gad8 (CBP‐Gad8). Eluates were loaded and immunoblotted (IB) using anti‐FLAG or anti‐CBP antibodies, together with 1% of whole cell extracts (WCE), to detect Taf12 co‐precipitation with Gad8. A strain which does not contain any TAP tag is shown as a negative control for the IP. Shown is an IB representative of two independent experiments.
Endogenous Gad8‐HA was affinity‐purified from starved cells, in order to activate its kinase activity (left and middle panels), and mixed with recombinant, purified GST‐Taf12, GST‐Taf12‐5A, or GST‐Fkh2 proteins (right panels) in a kinase assay buffer. Each assay was then analyzed by IB and probed with an anti‐phospho‐AKT substrate antibody.
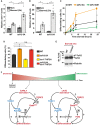
- A, B
Expression of ste11 + (A) and mei2 + (B) using quantitative RT–PCR of RNA extracted from cells grown either in rich medium (dark gray) or starved for 2 h (light gray). Cells of the following genotypes were analyzed: wild‐type isogenic controls (WT) and taf12‐5A mutants. act1 + served as a control for normalization across samples. Values from a WT strain grown in rich medium were set at 1 to allow comparisons across culture conditions and mutant strains. Each column represents the mean value of six independent experiments, overlaid with individual data points and SEM. Statistical significance was determined by two‐way ANOVA followed by Bonferroni's multiple comparison tests (n = 6); *P ≤ 0.01.
- C, D
Zygotes and tetrads, which correspond to differentiated cells, were counted from cultures of homothallic cells grown to mid‐log phase in rich medium (t0) and shifted to starvation medium for up to 24 h (C) or from heterothallic cells mixed and incubated for 2 days on mating medium (D). Cells of the following genotypes were analyzed: wild‐type isogenic controls (WT), taf12‐5A, taf12‐5DE, tor1‐T1972A, and taf12‐5A tor1‐T1972A. Each value represents the mean percentage and SEM of differentiating cells to the total number of cells, averaged from six independent experiments. At least 200 cells from the indicated genotypes were counted in each experiment. Statistical significance was determined by two‐way ANOVA followed by Tukey's multiple comparison tests (n = 6); *P ≤ 0.01.
- E
P‐Taf12 was followed by anti‐FLAG IB of protein extracts from WT and tor1‐T1972A strains, grown in rich medium and shifted to starved conditions for 60 min. An anti‐tubulin IB is shown as a control for loading. Shown is an IB which is representative of two independent experiments.
- F
Proposed model for the regulation and role of Taf12 phosphorylation during the early and late steps following nutrient starvation in S. pombe. Question marks represent unknown regulatory pathways. See Discussion for details.
Similar articles
-
A PP2A-B55-Mediated Crosstalk between TORC1 and TORC2 Regulates the Differentiation Response in Fission Yeast.Curr Biol. 2017 Jan 23;27(2):175-188. doi: 10.1016/j.cub.2016.11.037. Epub 2016 Dec 29. Curr Biol. 2017. PMID: 28041796 Free PMC article.
-
TOR complex 2 in fission yeast is required for chromatin-mediated gene silencing and assembly of heterochromatic domains at subtelomeres.J Biol Chem. 2018 May 25;293(21):8138-8150. doi: 10.1074/jbc.RA118.002270. Epub 2018 Apr 9. J Biol Chem. 2018. PMID: 29632066 Free PMC article.
-
Gad8 Protein Is Found in the Nucleus Where It Interacts with the MluI Cell Cycle Box-binding Factor (MBF) Transcriptional Complex to Regulate the Response to DNA Replication Stress.J Biol Chem. 2016 Apr 22;291(17):9371-81. doi: 10.1074/jbc.M115.705251. Epub 2016 Feb 24. J Biol Chem. 2016. PMID: 26912660 Free PMC article.
-
Fission Yeast TORC2 Signaling Pathway Ensures Cell Proliferation under Glucose-Limited, Nitrogen-Replete Conditions.Biomolecules. 2021 Oct 6;11(10):1465. doi: 10.3390/biom11101465. Biomolecules. 2021. PMID: 34680098 Free PMC article. Review.
-
TORC1-Dependent Phosphorylation Targets in Fission Yeast.Biomolecules. 2017 Jul 3;7(3):50. doi: 10.3390/biom7030050. Biomolecules. 2017. PMID: 28671615 Free PMC article. Review.
Cited by
-
TOR complex 2 contributes to regulation of gene expression via inhibiting Gcn5 recruitment to subtelomeric and DNA replication stress genes.PLoS Genet. 2022 Feb 14;18(2):e1010061. doi: 10.1371/journal.pgen.1010061. eCollection 2022 Feb. PLoS Genet. 2022. PMID: 35157728 Free PMC article.
-
Epigenetic Regulation of Autophagy Beyond the Cytoplasm: A Review.Front Cell Dev Biol. 2021 Jun 14;9:675599. doi: 10.3389/fcell.2021.675599. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34195194 Free PMC article. Review.
-
Greatwall-Endosulfine: A Molecular Switch that Regulates PP2A/B55 Protein Phosphatase Activity in Dividing and Quiescent Cells.Int J Mol Sci. 2019 Dec 10;20(24):6228. doi: 10.3390/ijms20246228. Int J Mol Sci. 2019. PMID: 31835586 Free PMC article. Review.
-
UPS writes a new saga of SAGA.Biochim Biophys Acta Gene Regul Mech. 2023 Dec;1866(4):194981. doi: 10.1016/j.bbagrm.2023.194981. Epub 2023 Aug 30. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 37657588 Free PMC article. Review.
-
Nuclear Functions of TOR: Impact on Transcription and the Epigenome.Genes (Basel). 2020 Jun 10;11(6):641. doi: 10.3390/genes11060641. Genes (Basel). 2020. PMID: 32532005 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

