Longitudinal genomic surveillance of MRSA in the UK reveals transmission patterns in hospitals and the community
- PMID: 29070701
- PMCID: PMC5683347
- DOI: 10.1126/scitranslmed.aak9745
Longitudinal genomic surveillance of MRSA in the UK reveals transmission patterns in hospitals and the community
Abstract
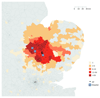
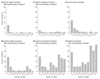
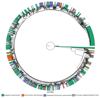
Genome sequencing has provided snapshots of the transmission of methicillin-resistant Staphylococcus aureus (MRSA) during suspected outbreaks in isolated hospital wards. Scale-up to populations is now required to establish the full potential of this technology for surveillance. We prospectively identified all individuals over a 12-month period who had at least one MRSA-positive sample processed by a routine diagnostic microbiology laboratory in the East of England, which received samples from three hospitals and 75 general practitioner (GP) practices. We sequenced at least 1 MRSA isolate from 1465 individuals (2282 MRSA isolates) and recorded epidemiological data. An integrated epidemiological and phylogenetic analysis revealed 173 transmission clusters containing between 2 and 44 cases and involving 598 people (40.8%). Of these, 118 clusters (371 people) involved hospital contacts alone, 27 clusters (72 people) involved community contacts alone, and 28 clusters (157 people) had both types of contact. Community- and hospital-associated MRSA lineages were equally capable of transmission in the community, with instances of spread in households, long-term care facilities, and GP practices. Our study provides a comprehensive picture of MRSA transmission in a sampled population of 1465 people and suggests the need to review existing infection control policy and practice.
Copyright © 2017 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures

Similar articles
-
Quantifying the transmission dynamics of MRSA in the community and healthcare settings in a low-prevalence country.Proc Natl Acad Sci U S A. 2019 Jul 16;116(29):14599-14605. doi: 10.1073/pnas.1900959116. Epub 2019 Jul 1. Proc Natl Acad Sci U S A. 2019. PMID: 31262808 Free PMC article.
-
Investigation of a Cluster of Sequence Type 22 Methicillin-Resistant Staphylococcus aureus Transmission in a Community Setting.Clin Infect Dis. 2017 Nov 29;65(12):2069-2077. doi: 10.1093/cid/cix539. Clin Infect Dis. 2017. PMID: 29077854 Free PMC article.
-
Emergence of SCCmec type IV and SCCmec type V methicillin-resistant Staphylococcus aureus containing the Panton-Valentine leukocidin genes in a large academic teaching hospital in central Switzerland: external invaders or persisting circulators?J Clin Microbiol. 2010 Mar;48(3):720-7. doi: 10.1128/JCM.01890-09. Epub 2009 Dec 30. J Clin Microbiol. 2010. PMID: 20042625 Free PMC article.
-
Community-associated meticillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated infection.J Hosp Infect. 2011 Nov;79(3):189-93. doi: 10.1016/j.jhin.2011.04.028. Epub 2011 Jul 7. J Hosp Infect. 2011. PMID: 21741111 Review.
-
Transmission and epidemiology of MRSA: current perspectives.Br J Nurs. 2005 May 26-Jun 8;14(10):548-51, 554. doi: 10.12968/bjon.2005.14.10.18103. Br J Nurs. 2005. PMID: 15928571 Review.
Cited by
-
Persisting uropathogenic Escherichia coli lineages show signatures of niche-specific within-host adaptation mediated by mobile genetic elements.Cell Host Microbe. 2022 Jul 13;30(7):1034-1047.e6. doi: 10.1016/j.chom.2022.04.008. Epub 2022 May 10. Cell Host Microbe. 2022. PMID: 35545083 Free PMC article.
-
Quantifying the transmission dynamics of MRSA in the community and healthcare settings in a low-prevalence country.Proc Natl Acad Sci U S A. 2019 Jul 16;116(29):14599-14605. doi: 10.1073/pnas.1900959116. Epub 2019 Jul 1. Proc Natl Acad Sci U S A. 2019. PMID: 31262808 Free PMC article.
-
Platanus_B: an accurate de novo assembler for bacterial genomes using an iterative error-removal process.DNA Res. 2020 Jun 1;27(3):dsaa014. doi: 10.1093/dnares/dsaa014. DNA Res. 2020. PMID: 32658266 Free PMC article.
-
Challenges and Opportunities for Global Genomic Surveillance Strategies in the COVID-19 Era.Viruses. 2022 Nov 16;14(11):2532. doi: 10.3390/v14112532. Viruses. 2022. PMID: 36423141 Free PMC article. Review.
-
Emerging variants of methicillin-resistant Staphylococcus aureus genotypes in Kuwait hospitals.PLoS One. 2018 Apr 18;13(4):e0195933. doi: 10.1371/journal.pone.0195933. eCollection 2018. PLoS One. 2018. PMID: 29668723 Free PMC article.
References
-
- Lowy FD. Staphylococcus aureus Infections. N Engl J Med. 1998;339:520–532. - PubMed
-
- Yaw LK, Robinson JO, Ho KM. A comparison of long-term outcomes after meticillin-resistant and meticillin-sensitive Staphylococcus aureus bacteraemia: an observational cohort study. Lancet Infect Dis. 2014;14:967–975. - PubMed
-
- Harris SR, Cartwright EJ, Török ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail Ma, Bentley SD, Parkhill J, Peacock SJ. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis. 2013;13:130–136. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

