Comparison of Whole Body SOD1 Knockout with Muscle-Specific SOD1 Knockout Mice Reveals a Role for Nerve Redox Signaling in Regulation of Degenerative Pathways in Skeletal Muscle
- PMID: 29065712
- PMCID: PMC5743036
- DOI: 10.1089/ars.2017.7249
Comparison of Whole Body SOD1 Knockout with Muscle-Specific SOD1 Knockout Mice Reveals a Role for Nerve Redox Signaling in Regulation of Degenerative Pathways in Skeletal Muscle
Abstract
Aims: Lack of Cu,Zn-superoxide dismutase (CuZnSOD) in homozygous knockout mice (Sod1-/-) leads to accelerated age-related muscle loss and weakness, but specific deletion of CuZnSOD in skeletal muscle (mSod1KO mice) or neurons (nSod1KO mice) resulted in only mild muscle functional deficits and failed to recapitulate the loss of mass and function observed in Sod1-/- mice. To dissect any underlying cross-talk between motor neurons and skeletal muscle in the degeneration in Sod1-/- mice, we characterized neuromuscular changes in the Sod1-/- model compared with mSod1KO mice and examined degenerative molecular mechanisms and pathways in peripheral nerve and skeletal muscle.
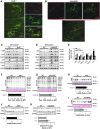
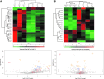
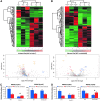
Results: In contrast to mSod1KO mice, myofiber atrophy in Sod1-/- mice was associated with increased muscle oxidative damage, neuromuscular junction degeneration, denervation, nerve demyelination, and upregulation of proteins involved in maintenance of myelin sheaths. Proteomic analyses confirmed increased proteasomal activity and adaptive stress responses in muscle of Sod1-/- mice that were absent in mSod1KO mice. Peripheral nerve from neither Sod1-/- nor mSod1KO mice showed increased oxidative damage or molecular responses to increased oxidation compared with wild type mice. Differential cysteine (Cys) labeling revealed a specific redox shift in the catalytic Cys residue of peroxiredoxin 6 (Cys47) in the peripheral nerve from Sod1-/- mice. Innovation and Conclusion: These findings demonstrate that neuromuscular integrity, redox mechanisms, and pathways are differentially altered in nerve and muscle of Sod1-/- and mSod1KO mice. Results support the concept that impaired redox signaling, rather than oxidative damage, in peripheral nerve plays a key role in muscle loss in Sod1-/- mice and potentially sarcopenia during aging. Antioxid. Redox Signal. 28, 275-295.
Keywords: 20S proteasome; mitochondria; myelin; peroxiredoxins 5 and 6; superoxide.
Conflict of interest statement
No competing financial interests exist.
Figures




Similar articles
-
Accelerated sarcopenia in Cu/Zn superoxide dismutase knockout mice.Free Radic Biol Med. 2019 Feb 20;132:19-23. doi: 10.1016/j.freeradbiomed.2018.06.032. Epub 2018 Jul 2. Free Radic Biol Med. 2019. PMID: 30670156 Free PMC article. Review.
-
Neuron-specific expression of CuZnSOD prevents the loss of muscle mass and function that occurs in homozygous CuZnSOD-knockout mice.FASEB J. 2014 Apr;28(4):1666-81. doi: 10.1096/fj.13-240390. Epub 2013 Dec 30. FASEB J. 2014. PMID: 24378874 Free PMC article.
-
Neuron specific reduction in CuZnSOD is not sufficient to initiate a full sarcopenia phenotype.Redox Biol. 2015 Aug;5:140-148. doi: 10.1016/j.redox.2015.04.005. Epub 2015 Apr 15. Redox Biol. 2015. PMID: 25917273 Free PMC article.
-
Ageing-induced changes in the redox status of peripheral motor nerves imply an effect on redox signalling rather than oxidative damage.Free Radic Biol Med. 2016 May;94:27-35. doi: 10.1016/j.freeradbiomed.2016.02.008. Epub 2016 Feb 10. Free Radic Biol Med. 2016. PMID: 26876649 Free PMC article.
-
Redox homeostasis and age-related deficits in neuromuscular integrity and function.J Cachexia Sarcopenia Muscle. 2017 Dec;8(6):881-906. doi: 10.1002/jcsm.12223. Epub 2017 Jul 26. J Cachexia Sarcopenia Muscle. 2017. PMID: 28744984 Free PMC article. Review.
Cited by
-
From the Bench to the Bedside: Branched Amino Acid and Micronutrient Strategies to Improve Mitochondrial Dysfunction Leading to Sarcopenia.Nutrients. 2022 Jan 22;14(3):483. doi: 10.3390/nu14030483. Nutrients. 2022. PMID: 35276842 Free PMC article. Review.
-
Interplay between Protein Kinase C Epsilon and Reactive Oxygen Species during Myogenic Differentiation.Cells. 2023 Jul 5;12(13):1792. doi: 10.3390/cells12131792. Cells. 2023. PMID: 37443826 Free PMC article.
-
Examination of SOD1 aggregation modulators and their effect on SOD1 enzymatic activity as a proxy for potential toxicity.FASEB J. 2020 Sep;34(9):11957-11969. doi: 10.1096/fj.202000948R. Epub 2020 Jul 23. FASEB J. 2020. PMID: 32701214 Free PMC article.
-
A Single Transcript Knockdown-Replacement Strategy Employing 5' UTR Secondary Structures to Precisely Titrate Rescue Protein Translation.Front Genome Ed. 2022 Mar 28;4:803375. doi: 10.3389/fgeed.2022.803375. eCollection 2022. Front Genome Ed. 2022. PMID: 35419562 Free PMC article.
-
Accelerated sarcopenia in Cu/Zn superoxide dismutase knockout mice.Free Radic Biol Med. 2019 Feb 20;132:19-23. doi: 10.1016/j.freeradbiomed.2018.06.032. Epub 2018 Jul 2. Free Radic Biol Med. 2019. PMID: 30670156 Free PMC article. Review.
References
-
- Bhattacharya A, Hamilton R, Jernigan A, Zhang Y, Sabia M, Rahman MM, Li Y, Wei R, Chaudhuri A, and Van Remmen H. Genetic ablation of 12/15-lipoxygenase but not 5-lipoxygenase protects against denervation-induced muscle atrophy. Free Radic Biol Med 67: 30–40, 2014 - PubMed
-
- Bronstein JM. Function of tetraspan proteins in the myelin sheath. Curr Opin Neurobiol 10: 552–557, 2000 - PubMed
-
- Broome CS, Kayani AC, Palomero J, Dillmann WH, Mestril R, Jackson MJ, and McArdle A. Effect of lifelong overexpression of HSP70 in skeletal muscle on age-related oxidative stress and adaptation after nondamaging contractile activity. FASEB J 20: 1549–1551, 2006 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

