Energetics of Glutamate Binding to an Ionotropic Glutamate Receptor
- PMID: 29065265
- PMCID: PMC5716343
- DOI: 10.1021/acs.jpcb.7b06862
Energetics of Glutamate Binding to an Ionotropic Glutamate Receptor
Abstract
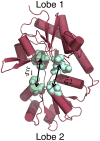
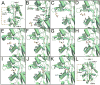
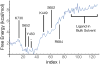
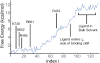
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that are responsible for the majority of excitatory transmission at the synaptic cleft. Mechanically speaking, agonist binding to the ligand binding domain (LBD) activates the receptor by triggering a conformational change that is transmitted to the transmembrane region, opening the ion channel pore. We use fully atomistic molecular dynamics simulations to investigate the binding process in the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor, an iGluR subtype. The string method with swarms of trajectories was applied to calculate the possible pathways glutamate traverses during ligand binding. Residues peripheral to the binding cleft are found to metastably bind the ligand prior to ligand entry into the binding pocket. Umbrella sampling simulations were performed to compute the free energy barriers along the binding pathways. The calculated free energy profiles demonstrate that metastable interactions contribute substantially to the energetics of ligand binding and form local minima in the overall free energy landscape. Protein-ligand interactions at sites outside of the orthosteric agonist-binding site may serve to lower the transition barriers of the binding process.
Figures

Similar articles
-
Neurotransmitter Funneling Optimizes Glutamate Receptor Kinetics.Neuron. 2018 Jan 3;97(1):139-149.e4. doi: 10.1016/j.neuron.2017.11.024. Epub 2017 Dec 14. Neuron. 2018. PMID: 29249286 Free PMC article.
-
Gating Motions and Stationary Gating Properties of Ionotropic Glutamate Receptors: Computation Meets Electrophysiology.Acc Chem Res. 2017 Apr 18;50(4):814-822. doi: 10.1021/acs.accounts.6b00598. Epub 2017 Feb 10. Acc Chem Res. 2017. PMID: 28186717 Free PMC article. Review.
-
Enhanced sampling of glutamate receptor ligand-binding domains.Neurosci Lett. 2019 May 1;700:17-21. doi: 10.1016/j.neulet.2018.04.018. Epub 2018 Apr 14. Neurosci Lett. 2019. PMID: 29665428 Review.
-
Computation of standard binding free energies of polar and charged ligands to the glutamate receptor GluA2.J Phys Chem B. 2014 Feb 20;118(7):1813-24. doi: 10.1021/jp412195m. Epub 2014 Feb 10. J Phys Chem B. 2014. PMID: 24479628
-
Full and partial agonism of ionotropic glutamate receptors indicated by molecular dynamics simulations.J Chem Inf Model. 2011 May 23;51(5):1037-47. doi: 10.1021/ci2000055. Epub 2011 May 2. J Chem Inf Model. 2011. PMID: 21500800
Cited by
-
Phosphorylation at Ser65 modulates ubiquitin conformational dynamics.Structure. 2023 Jul 6;31(7):884-890.e2. doi: 10.1016/j.str.2023.05.006. Epub 2023 Jun 1. Structure. 2023. PMID: 37267945 Free PMC article.
-
Sustained-release behavior and the antitumor effect of charge-convertible poly(amino acid)s drug-loaded nanoparticles.Drug Deliv Transl Res. 2023 Sep;13(9):2394-2406. doi: 10.1007/s13346-023-01323-w. Epub 2023 Mar 13. Drug Deliv Transl Res. 2023. PMID: 36913103
-
Activation and desensitization of ionotropic glutamate receptors by selectively triggering pre-existing motions.Neurosci Lett. 2019 May 1;700:22-29. doi: 10.1016/j.neulet.2018.02.050. Epub 2018 Feb 23. Neurosci Lett. 2019. PMID: 29481851 Free PMC article. Review.
-
Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain.Structure. 2019 Feb 5;27(2):241-252.e3. doi: 10.1016/j.str.2018.10.017. Epub 2018 Dec 6. Structure. 2019. PMID: 30528594 Free PMC article.
-
Atomic-scale characterization of mature HIV-1 capsid stabilization by inositol hexakisphosphate (IP6).Sci Adv. 2020 Sep 16;6(38):eabc6465. doi: 10.1126/sciadv.abc6465. Print 2020 Sep. Sci Adv. 2020. PMID: 32938668 Free PMC article.
References
-
- Mayer ML. Glutamate Receptors at Atomic Resolution. Nature. 2006;440:456–462. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

