Comparative Analysis of Tat-Dependent and Tat-Deficient Natural Lentiviruses
- PMID: 29061947
- PMCID: PMC5644649
- DOI: 10.3390/vetsci2040293
Comparative Analysis of Tat-Dependent and Tat-Deficient Natural Lentiviruses
Abstract
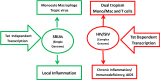
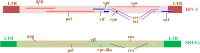
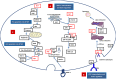
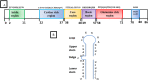
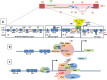
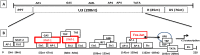
The emergence of human immunodeficiency virus (HIV) causing acquired immunodeficiency syndrome (AIDS) in infected humans has resulted in a global pandemic that has killed millions. HIV-1 and HIV-2 belong to the lentivirus genus of the Retroviridae family. This genus also includes viruses that infect other vertebrate animals, among them caprine arthritis-encephalitis virus (CAEV) and Maedi-Visna virus (MVV), the prototypes of a heterogeneous group of viruses known as small ruminant lentiviruses (SRLVs), affecting both goat and sheep worldwide. Despite their long host-SRLV natural history, SRLVs were never found to be responsible for immunodeficiency in contrast to primate lentiviruses. SRLVs only replicate productively in monocytes/macrophages in infected animals but not in CD4+ T cells. The focus of this review is to examine and compare the biological and pathological properties of SRLVs as prototypic Tat-independent lentiviruses with HIV-1 as prototypic Tat-dependent lentiviruses. Results from this analysis will help to improve the understanding of why and how these two prototypic lentiviruses evolved in opposite directions in term of virulence and pathogenicity. Results may also help develop new strategies based on the attenuation of SRLVs to control the highly pathogenic HIV-1 in humans.
Keywords: CAEV; HIV; Lentiviruses; Tat; latency; pathogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Small ruminant lentiviruses (SRLVs) break the species barrier to acquire new host range.Viruses. 2013 Jul 23;5(7):1867-84. doi: 10.3390/v5071867. Viruses. 2013. PMID: 23881276 Free PMC article. Review.
-
Genetic characterization of small ruminant lentiviruses circulating in naturally infected sheep and goats in Ontario, Canada.Virus Res. 2013 Jul;175(1):30-44. doi: 10.1016/j.virusres.2013.03.019. Epub 2013 Apr 10. Virus Res. 2013. PMID: 23583225
-
Lack of trans-activation function for Maedi Visna virus and Caprine arthritis encephalitis virus Tat proteins.Virology. 2003 Mar 15;307(2):317-27. doi: 10.1016/s0042-6822(02)00076-4. Virology. 2003. PMID: 12667801
-
Maedi-visna virus and its relationship to human immunodeficiency virus.AIDS Rev. 2005 Oct-Dec;7(4):233-45. AIDS Rev. 2005. PMID: 16425963 Review.
-
Maedi-visna virus and caprine arthritis encephalitis virus genomes encode a Vpr-like but no Tat protein.J Virol. 2003 Sep;77(17):9632-8. doi: 10.1128/jvi.77.17.9632-9638.2003. J Virol. 2003. PMID: 12915575 Free PMC article.
Cited by
-
The Vif protein of caprine arthritis encephalitis virus inhibits interferon production.Arch Virol. 2020 Jul;165(7):1557-1567. doi: 10.1007/s00705-020-04637-z. Epub 2020 Apr 30. Arch Virol. 2020. PMID: 32356187
-
Prospects in Innate Immune Responses as Potential Control Strategies against Non-Primate Lentiviruses.Viruses. 2018 Aug 17;10(8):435. doi: 10.3390/v10080435. Viruses. 2018. PMID: 30126090 Free PMC article. Review.
-
Y44A Mutation in the Acidic Domain of HIV-2 Tat Impairs Viral Reverse Transcription and LTR-Transactivation.Int J Mol Sci. 2020 Aug 17;21(16):5907. doi: 10.3390/ijms21165907. Int J Mol Sci. 2020. PMID: 32824587 Free PMC article.
References
-
- Peeters M., Fransen K., Delaporte E., Van den Haesevelde M., Gershy-Damet G.M., Kestens L., van der Groen G., Piot P. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS. 1992;6:447–451. doi: 10.1097/00002030-199205000-00002. - DOI - PubMed
-
- Barre-Sinoussi F., Chermann J., Rey F., Nugeyre M., Chamaret S., Gruest J., Dauguet C., Axler B.C., Vezinet- Brun F., Rouzioux C., et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

