Hemodynamic Forces Sculpt Developing Heart Valves through a KLF2-WNT9B Paracrine Signaling Axis
- PMID: 29056552
- PMCID: PMC5760194
- DOI: 10.1016/j.devcel.2017.09.023
Hemodynamic Forces Sculpt Developing Heart Valves through a KLF2-WNT9B Paracrine Signaling Axis
Abstract
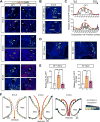
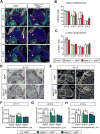
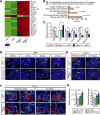
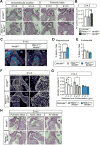
Hemodynamic forces play an essential epigenetic role in heart valve development, but how they do so is not known. Here, we show that the shear-responsive transcription factor KLF2 is required in endocardial cells to regulate the mesenchymal cell responses that remodel cardiac cushions to mature valves. Endocardial Klf2 deficiency results in defective valve formation associated with loss of Wnt9b expression and reduced canonical WNT signaling in neighboring mesenchymal cells, a phenotype reproduced by endocardial-specific loss of Wnt9b. Studies in zebrafish embryos reveal that wnt9b expression is similarly restricted to the endocardial cells overlying the developing heart valves and is dependent upon both hemodynamic shear forces and klf2a expression. These studies identify KLF2-WNT9B signaling as a conserved molecular mechanism by which fluid forces sensed by endothelial cells direct the complex cellular process of heart valve development and suggest that congenital valve defects may arise due to subtle defects in this mechanotransduction pathway.
Keywords: Klf2; Klf4; Wnt9b; cardiac cushion; endocardium; heart valve development; hemodynamic force.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Mechanosensitive Notch-Dll4 and Klf2-Wnt9 signaling pathways intersect in guiding valvulogenesis in zebrafish.Cell Rep. 2021 Oct 5;37(1):109782. doi: 10.1016/j.celrep.2021.109782. Cell Rep. 2021. PMID: 34610316 Free PMC article.
-
Oscillatory Flow Modulates Mechanosensitive klf2a Expression through trpv4 and trpp2 during Heart Valve Development.Curr Biol. 2015 May 18;25(10):1354-61. doi: 10.1016/j.cub.2015.03.038. Epub 2015 May 7. Curr Biol. 2015. PMID: 25959969
-
Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart.PLoS Biol. 2009 Nov;7(11):e1000246. doi: 10.1371/journal.pbio.1000246. Epub 2009 Nov 17. PLoS Biol. 2009. PMID: 19924233 Free PMC article.
-
Hemodynamics driven cardiac valve morphogenesis.Biochim Biophys Acta. 2016 Jul;1863(7 Pt B):1760-6. doi: 10.1016/j.bbamcr.2015.11.014. Epub 2015 Nov 30. Biochim Biophys Acta. 2016. PMID: 26608609 Review.
-
Cell biology of cardiac cushion development.Int Rev Cytol. 2005;243:287-335. doi: 10.1016/S0074-7696(05)43005-3. Int Rev Cytol. 2005. PMID: 15797462 Review.
Cited by
-
Drainage of amniotic fluid delays vocal fold separation and induces load-related vocal fold mucosa remodeling.Dev Biol. 2020 Oct 1;466(1-2):47-58. doi: 10.1016/j.ydbio.2020.08.003. Epub 2020 Aug 7. Dev Biol. 2020. PMID: 32777221 Free PMC article.
-
Multiscale structure and function of the aortic valve apparatus.Physiol Rev. 2024 Oct 1;104(4):1487-1532. doi: 10.1152/physrev.00038.2022. Epub 2023 Sep 21. Physiol Rev. 2024. PMID: 37732828 Free PMC article. Review.
-
Spatial transcriptomics reveals novel genes during the remodelling of the embryonic human arterial valves.PLoS Genet. 2023 Nov 27;19(11):e1010777. doi: 10.1371/journal.pgen.1010777. eCollection 2023 Nov. PLoS Genet. 2023. PMID: 38011284 Free PMC article.
-
Genetics of Cardiac Aging Implicate Organ-Specific Variation.medRxiv [Preprint]. 2024 Aug 6:2024.08.02.24310874. doi: 10.1101/2024.08.02.24310874. medRxiv. 2024. PMID: 39148824 Free PMC article. Preprint.
-
Loss of flow responsive Tie1 results in Impaired Aortic valve remodeling.Dev Biol. 2019 Nov 1;455(1):73-84. doi: 10.1016/j.ydbio.2019.07.011. Epub 2019 Jul 15. Dev Biol. 2019. PMID: 31319059 Free PMC article.
References
-
- Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. - PubMed
-
- Chang CP, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, Graef IA, Crabtree GR. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell. 2004;118:649–663. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

