The 'how' and 'where' of plant microRNAs
- PMID: 29048752
- PMCID: PMC6040672
- DOI: 10.1111/nph.14834
The 'how' and 'where' of plant microRNAs
Abstract
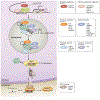
Contents 1002 I. 1002 II. 1007 III. 1010 IV. 1013 1013 References 1013 SUMMARY: MicroRNAs (miRNAs) are small non-coding RNAs, of typically 20-24 nt, that regulate gene expression post-transcriptionally through sequence complementarity. Since the identification of the first miRNA, lin-4, in the nematode Caenorhabditis elegans in 1993, thousands of miRNAs have been discovered in animals and plants, and their regulatory roles in numerous biological processes have been uncovered. In plants, research efforts have established the major molecular framework of miRNA biogenesis and modes of action, and are beginning to elucidate the mechanisms of miRNA degradation. Studies have implicated restricted and surprising subcellular locations in which miRNA biogenesis or activity takes place. In this article, we summarize the current knowledge on how plant miRNAs are made and degraded, and how they repress target gene expression. We discuss not only the players involved in these processes, but also the subcellular sites in which these processes are known or implicated to take place. We hope to raise awareness that the cell biology of miRNAs holds the key to a full understanding of these enigmatic molecules.
Keywords: ARGONAUTE1; DICER-LIKE1; HYPONASTIC LEAVES 1; dicing body; endoplasmic reticulum (ER); membrane-bound polysome; microRNA; phased small interfering RNA.
© 2017 The Authors. New Phytologist © 2017 New Phytologist Trust.
Figures


Similar articles
-
Recent advances in the regulation of plant miRNA biogenesis.RNA Biol. 2021 Dec;18(12):2087-2096. doi: 10.1080/15476286.2021.1899491. Epub 2021 Mar 17. RNA Biol. 2021. PMID: 33666136 Free PMC article. Review.
-
microRNA production in Arabidopsis.Front Plant Sci. 2023 Jan 19;14:1096772. doi: 10.3389/fpls.2023.1096772. eCollection 2023. Front Plant Sci. 2023. PMID: 36743500 Free PMC article. Review.
-
Functional characterization of a 'plant-like' HYL1 homolog in the cnidarian Nematostella vectensis indicates a conserved involvement in microRNA biogenesis.Elife. 2022 Mar 15;11:e69464. doi: 10.7554/eLife.69464. Elife. 2022. PMID: 35289745 Free PMC article.
-
microRNA biogenesis, degradation and activity in plants.Cell Mol Life Sci. 2015 Jan;72(1):87-99. doi: 10.1007/s00018-014-1728-7. Epub 2014 Sep 11. Cell Mol Life Sci. 2015. PMID: 25209320 Free PMC article. Review.
-
Plant microRNAs and development.J Genet Genomics. 2013 May 20;40(5):217-30. doi: 10.1016/j.jgg.2013.04.002. Epub 2013 Apr 15. J Genet Genomics. 2013. PMID: 23706297 Review.
Cited by
-
Assessment of heat tolerance and identification of miRNAs during high-temperature response in grapevine.Front Plant Sci. 2024 Oct 22;15:1484892. doi: 10.3389/fpls.2024.1484892. eCollection 2024. Front Plant Sci. 2024. PMID: 39502927 Free PMC article.
-
Gma-miR408 Enhances Soybean Cyst Nematode Susceptibility by Suppressing Reactive Oxygen Species Accumulation.Int J Mol Sci. 2022 Nov 14;23(22):14022. doi: 10.3390/ijms232214022. Int J Mol Sci. 2022. PMID: 36430501 Free PMC article.
-
Genome-wide analysis of plant miRNA action clarifies levels of regulatory dynamics across developmental contexts.Genome Res. 2021 May;31(5):811-822. doi: 10.1101/gr.270918.120. Epub 2021 Apr 16. Genome Res. 2021. PMID: 33863807 Free PMC article.
-
Tiny Yet Indispensable Plant MicroRNAs Are Worth to Explore as Key Components for Combating Genotoxic Stresses.Front Plant Sci. 2019 Oct 4;10:1197. doi: 10.3389/fpls.2019.01197. eCollection 2019. Front Plant Sci. 2019. PMID: 31636646 Free PMC article. Review.
-
Prevalent cytidylation and uridylation of precursor miRNAs in Arabidopsis.Nat Plants. 2019 Dec;5(12):1260-1272. doi: 10.1038/s41477-019-0562-1. Epub 2019 Dec 2. Nat Plants. 2019. PMID: 31792392
References
-
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

