Phosphorylation of iRhom2 Controls Stimulated Proteolytic Shedding by the Metalloprotease ADAM17/TACE
- PMID: 29045841
- PMCID: PMC5656746
- DOI: 10.1016/j.celrep.2017.09.074
Phosphorylation of iRhom2 Controls Stimulated Proteolytic Shedding by the Metalloprotease ADAM17/TACE
Abstract
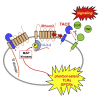
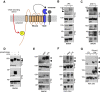
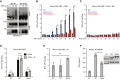
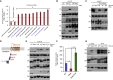
Cell surface metalloproteases coordinate signaling during development, tissue homeostasis, and disease. TACE (TNF-α-converting enzyme), is responsible for cleavage ("shedding") of membrane-tethered signaling molecules, including the cytokine TNF, and activating ligands of the EGFR. The trafficking of TACE within the secretory pathway requires its binding to iRhom2, which mediates the exit of TACE from the endoplasmic reticulum. An important, but mechanistically unclear, feature of TACE biology is its ability to be stimulated rapidly on the cell surface by numerous inflammatory and growth-promoting agents. Here, we report a role for iRhom2 in TACE stimulation on the cell surface. TACE shedding stimuli trigger MAP kinase-dependent phosphorylation of iRhom2 N-terminal cytoplasmic tail. This recruits 14-3-3 proteins, enforcing the dissociation of TACE from complexes with iRhom2, promoting the cleavage of TACE substrates. Our data reveal that iRhom2 controls multiple aspects of TACE biology, including stimulated shedding on the cell surface.
Keywords: 14-3-3; ADAM metalloproteases; ADAM17/TACE; EGFR; MAP kinases; TNF; ectodomain shedding; iRhom2.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Phosphorylation of iRhom2 at the plasma membrane controls mammalian TACE-dependent inflammatory and growth factor signalling.Elife. 2017 Apr 22;6:e23968. doi: 10.7554/eLife.23968. Elife. 2017. PMID: 28432785 Free PMC article.
-
Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE.Science. 2012 Jan 13;335(6065):225-8. doi: 10.1126/science.1214400. Science. 2012. PMID: 22246777 Free PMC article.
-
iRhom2 controls the substrate selectivity of stimulated ADAM17-dependent ectodomain shedding.Proc Natl Acad Sci U S A. 2013 Jul 9;110(28):11433-8. doi: 10.1073/pnas.1302553110. Epub 2013 Jun 25. Proc Natl Acad Sci U S A. 2013. PMID: 23801765 Free PMC article.
-
iRhom2: An Emerging Adaptor Regulating Immunity and Disease.Int J Mol Sci. 2020 Sep 8;21(18):6570. doi: 10.3390/ijms21186570. Int J Mol Sci. 2020. PMID: 32911849 Free PMC article. Review.
-
Mechanistic insight on the role of iRhom2-TNF-α-BAFF signaling pathway in various autoimmune disorders.Adv Biol Regul. 2024 May;92:101011. doi: 10.1016/j.jbior.2023.101011. Epub 2023 Dec 14. Adv Biol Regul. 2024. PMID: 38151421 Review.
Cited by
-
Innovations in Biomaterial Design toward Successful RNA Interference Therapy for Cancer Treatment.Adv Healthc Mater. 2021 Jul;10(13):e2100350. doi: 10.1002/adhm.202100350. Epub 2021 May 11. Adv Healthc Mater. 2021. PMID: 33973393 Free PMC article. Review.
-
Transmembrane TNF and Its Receptors TNFR1 and TNFR2 in Mycobacterial Infections.Int J Mol Sci. 2021 May 22;22(11):5461. doi: 10.3390/ijms22115461. Int J Mol Sci. 2021. PMID: 34067256 Free PMC article. Review.
-
Systematic analysis of the Frazzled receptor interactome establishes previously unreported regulators of axon guidance.Development. 2023 Aug 1;150(15):dev201636. doi: 10.1242/dev.201636. Epub 2023 Aug 1. Development. 2023. PMID: 37526651 Free PMC article.
-
iRhom2 regulates ERBB signalling to promote KRAS-driven tumour growth of lung cancer cells.J Cell Sci. 2022 Sep 1;135(17):jcs259949. doi: 10.1242/jcs.259949. Epub 2022 Sep 8. J Cell Sci. 2022. PMID: 35971826 Free PMC article.
-
C11orf94/Frey is a key regulator for male fertility by controlling Izumo1 complex assembly.Sci Adv. 2022 Aug 12;8(32):eabo6049. doi: 10.1126/sciadv.abo6049. Epub 2022 Aug 12. Sci Adv. 2022. PMID: 35960805 Free PMC article.
References
-
- Adrain C., Freeman M. New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat. Rev. Mol. Cell Biol. 2012;13:489–498. - PubMed
-
- Arribas J., Coodly L., Vollmer P., Kishimoto T.K., Rose-John S., Massagué J. Diverse cell surface protein ectodomains are shed by a system sensitive to metalloprotease inhibitors. J. Biol. Chem. 1996;271:11376–11382. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

