MNK1/2 inhibition limits oncogenicity and metastasis of KIT-mutant melanoma
- PMID: 29035277
- PMCID: PMC5663367
- DOI: 10.1172/JCI91258
MNK1/2 inhibition limits oncogenicity and metastasis of KIT-mutant melanoma
Erratum in
-
MNK1/2 inhibition limits oncogenicity and metastasis of KIT-mutant melanoma.J Clin Invest. 2024 Apr 15;134(8):e181338. doi: 10.1172/JCI181338. J Clin Invest. 2024. PMID: 38618965 Free PMC article. No abstract available.
Abstract
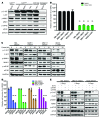
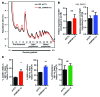
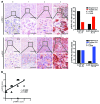
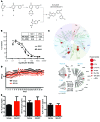
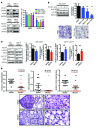
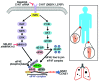
Melanoma can be stratified into unique subtypes based on distinct pathologies. The acral/mucosal melanoma subtype is characterized by aberrant and constitutive activation of the proto-oncogene receptor tyrosine kinase C-KIT, which drives tumorigenesis. Treatment of these melanoma patients with C-KIT inhibitors has proven challenging, prompting us to investigate the downstream effectors of the C-KIT receptor. We determined that C-KIT stimulates MAP kinase-interacting serine/threonine kinases 1 and 2 (MNK1/2), which phosphorylate eukaryotic translation initiation factor 4E (eIF4E) and render it oncogenic. Depletion of MNK1/2 in melanoma cells with oncogenic C-KIT inhibited cell migration and mRNA translation of the transcriptional repressor SNAI1 and the cell cycle gene CCNE1. This suggested that blocking MNK1/2 activity may inhibit tumor progression, at least in part, by blocking translation initiation of mRNAs encoding cell migration proteins. Moreover, we developed an MNK1/2 inhibitor (SEL201), and found that SEL201-treated KIT-mutant melanoma cells had lower oncogenicity and reduced metastatic ability. Clinically, tumors from melanoma patients harboring KIT mutations displayed a marked increase in MNK1 and phospho-eIF4E. Thus, our studies indicate that blocking MNK1/2 exerts potent antimelanoma effects and support blocking MNK1/2 as a potential strategy to treat patients positive for KIT mutations.
Conflict of interest statement
Figures

Similar articles
-
MNK1 signaling induces an ANGPTL4-mediated gene signature to drive melanoma progression.Oncogene. 2020 Apr;39(18):3650-3665. doi: 10.1038/s41388-020-1240-5. Epub 2020 Mar 4. Oncogene. 2020. PMID: 32132651
-
CGP57380 enhances efficacy of RAD001 in non-small cell lung cancer through abrogating mTOR inhibition-induced phosphorylation of eIF4E and activating mitochondrial apoptotic pathway.Oncotarget. 2016 May 10;7(19):27787-801. doi: 10.18632/oncotarget.8497. Oncotarget. 2016. PMID: 27050281 Free PMC article.
-
Design, synthesis and biological evaluation of pyridone-aminal derivatives as MNK1/2 inhibitors.Bioorg Med Chem. 2019 Apr 1;27(7):1211-1225. doi: 10.1016/j.bmc.2019.02.007. Epub 2019 Feb 2. Bioorg Med Chem. 2019. PMID: 30824167
-
The MNK1/2-eIF4E Axis as a Potential Therapeutic Target in Melanoma.Int J Mol Sci. 2020 Jun 5;21(11):4055. doi: 10.3390/ijms21114055. Int J Mol Sci. 2020. PMID: 32517051 Free PMC article. Review.
-
Mitogen-activated Protein Kinase (MAPK) Interacting Kinases 1 and 2 (MNK1 and MNK2) as Targets for Cancer Therapy: Recent Progress in the Development of MNK Inhibitors.Curr Med Chem. 2017;24(28):3025-3053. doi: 10.2174/0929867324666170203123427. Curr Med Chem. 2017. PMID: 28164761 Review.
Cited by
-
Inhibition of MNKs promotes macrophage immunosuppressive phenotype to limit CD8+ T cell antitumor immunity.JCI Insight. 2022 May 9;7(9):e152731. doi: 10.1172/jci.insight.152731. JCI Insight. 2022. PMID: 35380995 Free PMC article.
-
Translation regulation in skin cancer from a tRNA point of view.Epigenomics. 2019 Feb;11(2):215-245. doi: 10.2217/epi-2018-0176. Epub 2018 Dec 19. Epigenomics. 2019. PMID: 30565492 Free PMC article. Review.
-
Repurposing Ponatinib as a Potent Agent against KIT Mutant Melanomas.Theranostics. 2019 Mar 16;9(7):1952-1964. doi: 10.7150/thno.30890. eCollection 2019. Theranostics. 2019. PMID: 31037149 Free PMC article.
-
Cooperative activation of PDK1 and AKT by MAPK4 enhances cancer growth and resistance to therapy.PLoS Biol. 2023 Aug 2;21(8):e3002227. doi: 10.1371/journal.pbio.3002227. eCollection 2023 Aug. PLoS Biol. 2023. PMID: 37531320 Free PMC article.
-
Deeping in the Role of the MAP-Kinases Interacting Kinases (MNKs) in Cancer.Int J Mol Sci. 2020 Apr 23;21(8):2967. doi: 10.3390/ijms21082967. Int J Mol Sci. 2020. PMID: 32340135 Free PMC article. Review.
References
-
- Chan KK, Chan RC, Ho RS, Chan JY. Clinical patterns of melanoma in Asians: 11-year experience in a tertiary referral center. Ann Plast Surg. 2016;77(suppl 1):S6–S11. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

