Dysregulation of the Mitochondrial Unfolded Protein Response Induces Non-Apoptotic Dopaminergic Neurodegeneration in C. elegans Models of Parkinson's Disease
- PMID: 29030433
- PMCID: PMC5688521
- DOI: 10.1523/JNEUROSCI.1294-17.2017
Dysregulation of the Mitochondrial Unfolded Protein Response Induces Non-Apoptotic Dopaminergic Neurodegeneration in C. elegans Models of Parkinson's Disease
Abstract
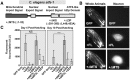
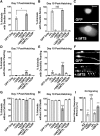
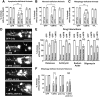
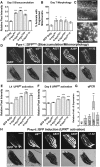
Due to environmental insult or innate genetic deficiency, protein folding environments of the mitochondrial matrix are prone to dysregulation, prompting the activation of a specific organellar stress-response mechanism, the mitochondrial unfolded protein response (UPRMT). In Caenorhabditis elegans, mitochondrial damage leads to nuclear translocation of the ATFS-1 transcription factor to activate the UPRMT After short-term acute stress has been mitigated, the UPRMT is eventually suppressed to restore homeostasis to C. elegans hermaphrodites. In contrast, and reflective of the more chronic nature of progressive neurodegenerative disorders such as Parkinson's disease (PD), here, we report the consequences of prolonged, cell-autonomous activation of the UPRMT in C. elegans dopaminergic neurons. We reveal that neuronal function and integrity decline rapidly with age, culminating in activity-dependent, non-apoptotic cell death. In a PD-like context wherein transgenic nematodes express the Lewy body constituent protein α-synuclein (αS), we not only find that this protein and its PD-associated disease variants have the capacity to induce the UPRMT, but also that coexpression of αS and ATFS-1-associated dysregulation of the UPRMT synergistically potentiate dopaminergic neurotoxicity. This genetic interaction is in parallel to mitophagic pathways dependent on the C. elegans PINK1 homolog, which is necessary for cellular resistance to chronic malfunction of the UPRMT Given the increasingly recognized role of mitochondrial quality control in neurodegenerative diseases, these studies illustrate, for the first time, an insidious aspect of mitochondrial signaling in which the UPRMT pathway, under disease-associated, context-specific dysregulation, exacerbates disruption of dopaminergic neurons in vivo, resulting in the neurodegeneration characteristic of PD.SIGNIFICANCE STATEMENT Disruptions or alterations in the activation of pathways that regulate mitochondrial quality control have been linked to neurodegenerative diseases due in part to the central role of mitochondria in metabolism, ROS regulation, and proteostasis. The extent to which these pathways, including the mitochondrial unfolded protein response (UPRMT) and mitophagy, are active may predict severity and progression of these disorders, as well as sensitivity to compounding stressors. Furthermore, therapeutic strategies that aim to induce these pathways may benefit from increased study into cellular responses that arise from long-term or ectopic stimulation, especially in neuronal compartments. By demonstrating the detrimental consequences of prolonged cellular activation of the UPRMT, we provide evidence that this pathway is not a universally beneficial mechanism because dysregulation has neurotoxic consequences.
Keywords: C. elegans; alpha-synuclein; dopaminergic; mitochondria; neurodegeneration; stress.
Copyright © 2017 the authors 0270-6474/17/3711085-16$15.00/0.
Figures
Similar articles
-
Sphingosine Kinase Activates the Mitochondrial Unfolded Protein Response and Is Targeted to Mitochondria by Stress.Cell Rep. 2018 Sep 11;24(11):2932-2945.e4. doi: 10.1016/j.celrep.2018.08.037. Cell Rep. 2018. PMID: 30208318 Free PMC article.
-
Genetic modulators associated with regulatory surveillance of mitochondrial quality control, play a key role in regulating stress pathways and longevity in C. elegans.Life Sci. 2022 Feb 1;290:120226. doi: 10.1016/j.lfs.2021.120226. Epub 2021 Dec 23. Life Sci. 2022. PMID: 34953889
-
Activation of the mitochondrial unfolded protein response promotes longevity and dopamine neuron survival in Parkinson's disease models.Sci Rep. 2017 Nov 27;7(1):16441. doi: 10.1038/s41598-017-16637-2. Sci Rep. 2017. PMID: 29180793 Free PMC article.
-
Sirtuins and the Estrogen Receptor as Regulators of the Mammalian Mitochondrial UPR in Cancer and Aging.Adv Cancer Res. 2016;130:211-56. doi: 10.1016/bs.acr.2016.01.004. Epub 2016 Mar 2. Adv Cancer Res. 2016. PMID: 27037754 Review.
-
The mitochondrial unfolded protein response: A multitasking giant in the fight against human diseases.Ageing Res Rev. 2022 Nov;81:101702. doi: 10.1016/j.arr.2022.101702. Epub 2022 Jul 28. Ageing Res Rev. 2022. PMID: 35908669 Review.
Cited by
-
Curcumin prevents Alzheimer's disease progression by upregulating JMJD3.Am J Transl Res. 2022 Aug 15;14(8):5280-5294. eCollection 2022. Am J Transl Res. 2022. PMID: 36105064 Free PMC article.
-
The Mitochondrial Unfolded Protein Response: A Hinge Between Healthy and Pathological Aging.Front Aging Neurosci. 2020 Sep 11;12:581849. doi: 10.3389/fnagi.2020.581849. eCollection 2020. Front Aging Neurosci. 2020. PMID: 33061907 Free PMC article. Review.
-
Interplay between Mitochondrial Protein Import and Respiratory Complexes Assembly in Neuronal Health and Degeneration.Life (Basel). 2021 May 11;11(5):432. doi: 10.3390/life11050432. Life (Basel). 2021. PMID: 34064758 Free PMC article. Review.
-
RNA-Binding Proteins Implicated in Mitochondrial Damage and Mitophagy.Front Cell Dev Biol. 2020 Jun 4;8:372. doi: 10.3389/fcell.2020.00372. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32582692 Free PMC article. Review.
-
Advancements in Genetic and Biochemical Insights: Unraveling the Etiopathogenesis of Neurodegeneration in Parkinson's Disease.Biomolecules. 2024 Jan 5;14(1):73. doi: 10.3390/biom14010073. Biomolecules. 2024. PMID: 38254673 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
