Interplay of pathogenic forms of human tau with different autophagic pathways
- PMID: 29024336
- PMCID: PMC5770880
- DOI: 10.1111/acel.12692
Interplay of pathogenic forms of human tau with different autophagic pathways
Abstract
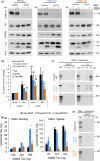
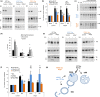
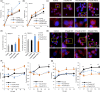
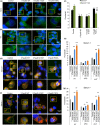
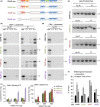
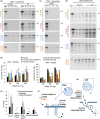
Loss of neuronal proteostasis, a common feature of the aging brain, is accelerated in neurodegenerative disorders, including different types of tauopathies. Aberrant turnover of tau, a microtubule-stabilizing protein, contributes to its accumulation and subsequent toxicity in tauopathy patients' brains. A direct toxic effect of pathogenic forms of tau on the proteolytic systems that normally contribute to their turnover has been proposed. In this study, we analyzed the contribution of three different types of autophagy, macroautophagy, chaperone-mediated autophagy, and endosomal microautophagy to the degradation of tau protein variants and tau mutations associated with this age-related disease. We have found that the pathogenic P301L mutation inhibits degradation of tau by any of the three autophagic pathways, whereas the risk-associated tau mutation A152T reroutes tau for degradation through a different autophagy pathway. We also found defective autophagic degradation of tau when using mutations that mimic common posttranslational modifications in tau or known to promote its aggregation. Interestingly, although most mutations markedly reduced degradation of tau through autophagy, the step of this process preferentially affected varies depending on the type of tau mutation. Overall, our studies unveil a complex interplay between the multiple modifications of tau and selective forms of autophagy that may determine its physiological degradation and its faulty clearance in the disease context.
Keywords: Alzheimer's disease; aging; autophagy; frontotemporal dementia; lysosomes; neurodegeneration.
© 2017 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures

Similar articles
-
Acetylated tau inhibits chaperone-mediated autophagy and promotes tau pathology propagation in mice.Nat Commun. 2021 Apr 14;12(1):2238. doi: 10.1038/s41467-021-22501-9. Nat Commun. 2021. PMID: 33854069 Free PMC article.
-
BAG3 and SYNPO (synaptopodin) facilitate phospho-MAPT/Tau degradation via autophagy in neuronal processes.Autophagy. 2019 Jul;15(7):1199-1213. doi: 10.1080/15548627.2019.1580096. Epub 2019 Mar 1. Autophagy. 2019. PMID: 30744518 Free PMC article.
-
Temsirolimus attenuates tauopathy in vitro and in vivo by targeting tau hyperphosphorylation and autophagic clearance.Neuropharmacology. 2014 Oct;85:121-30. doi: 10.1016/j.neuropharm.2014.05.032. Epub 2014 May 29. Neuropharmacology. 2014. PMID: 24880087
-
Tauopathies and tau oligomers.J Alzheimers Dis. 2013;37(3):565-8. doi: 10.3233/JAD-130653. J Alzheimers Dis. 2013. PMID: 23948895 Review.
-
Synergy and antagonism of macroautophagy and chaperone-mediated autophagy in a cell model of pathological tau aggregation.Autophagy. 2010 Jan;6(1):182-3. doi: 10.4161/auto.6.1.10815. Epub 2010 Jan 1. Autophagy. 2010. PMID: 20023429 Review.
Cited by
-
Age-dependent accumulation of tau aggregation in Caenorhabditis elegans.Front Aging. 2022 Aug 19;3:928574. doi: 10.3389/fragi.2022.928574. eCollection 2022. Front Aging. 2022. PMID: 36062211 Free PMC article.
-
Dysregulated proteostasis network in neuronal diseases.Front Cell Dev Biol. 2023 Feb 24;11:1075215. doi: 10.3389/fcell.2023.1075215. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36910151 Free PMC article. Review.
-
Protein acylation: mechanisms, biological functions and therapeutic targets.Signal Transduct Target Ther. 2022 Dec 29;7(1):396. doi: 10.1038/s41392-022-01245-y. Signal Transduct Target Ther. 2022. PMID: 36577755 Free PMC article. Review.
-
Oxidative Stress in Tauopathies: From Cause to Therapy.Antioxidants (Basel). 2022 Jul 22;11(8):1421. doi: 10.3390/antiox11081421. Antioxidants (Basel). 2022. PMID: 35892623 Free PMC article. Review.
-
Tau and other proteins found in Alzheimer's disease spinal fluid are linked to retromer-mediated endosomal traffic in mice and humans.Sci Transl Med. 2020 Nov 25;12(571):eaba6334. doi: 10.1126/scitranslmed.aba6334. Sci Transl Med. 2020. PMID: 33239387 Free PMC article.
References
-
- Aniento F, Roche E, Cuervo AM, Knecht E (1993) Uptake and degradation of glyceraldehyde‐3‐ phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem. 268, 10463–10470. - PubMed
-
- Barghorn S, Mandelkow E (2002) Toward a unified scheme for the aggregation of tau into Alzheimer paired helical filaments. Biochemistry 41, 14885–14896. - PubMed
-
- Barghorn S, Zheng‐Fischhofer Q, Ackmann M, Biernat J, von Bergen M, Mandelkow EM, Mandelkow E (2000) Structure, microtubule interactions, and paired helical filament aggregation by tau mutants of frontotemporal dementias. Biochemistry 39, 11714–11721. - PubMed
-
- Barghorn S, Biernat J, Mandelkow E (2005) Purification of recombinant tau protein and preparation of Alzheimer‐paired helical filaments in vitro . Methods Mol. Biol. 299, 35–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

