Inspirations on Virus Replication and Cell-to-Cell Movement from Studies Examining the Cytopathology Induced by Lettuce infectious yellows virus in Plant Cells
- PMID: 29021801
- PMCID: PMC5623981
- DOI: 10.3389/fpls.2017.01672
Inspirations on Virus Replication and Cell-to-Cell Movement from Studies Examining the Cytopathology Induced by Lettuce infectious yellows virus in Plant Cells
Abstract
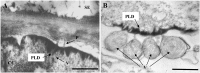
Lettuce infectious yellows virus (LIYV) is the type member of the genus Crinivirus in the family Closteroviridae. Like many other positive-strand RNA viruses, LIYV infections induce a number of cytopathic changes in plant cells, of which the two most characteristic are: Beet yellows virus-type inclusion bodies composed of vesicles derived from cytoplasmic membranes; and conical plasmalemma deposits (PLDs) located at the plasmalemma over plasmodesmata pit fields. The former are not only found in various closterovirus infections, but similar structures are known as 'viral factories' or viroplasms in cells infected with diverse types of animal and plant viruses. These are generally sites of virus replication, virion assembly and in some cases are involved in cell-to-cell transport. By contrast, PLDs induced by the LIYV-encoded P26 non-virion protein are not involved in replication but are speculated to have roles in virus intercellular movement. These deposits often harbor LIYV virions arranged to be perpendicular to the plasma membrane over plasmodesmata, and our recent studies show that P26 is required for LIYV systemic plant infection. The functional mechanism of how LIYV P26 facilitates intercellular movement remains unclear, however, research on other plant viruses provides some insights on the possible ways of viral intercellular movement through targeting and modifying plasmodesmata via interactions between plant cellular components and viral-encoded factors. In summary, beginning with LIYV, we review the studies that have uncovered the biological determinants giving rise to these cytopathological effects and their importance in viral replication, virion assembly and intercellular movement during the plant infection by closteroviruses, and compare these findings with those for other positive-strand RNA viruses.
Keywords: Closteroviridae; cytopathology; intercellular movement; membrane remodeling; plasmodesmata; virus replication.
Figures


Similar articles
-
A Distinct, Non-Virion Plant Virus Movement Protein Encoded by a Crinivirus Essential for Systemic Infection.mBio. 2018 Nov 20;9(6):e02230-18. doi: 10.1128/mBio.02230-18. mBio. 2018. PMID: 30459200 Free PMC article.
-
Lettuce infectious yellows virus-encoded P26 induces plasmalemma deposit cytopathology.Virology. 2009 May 25;388(1):212-20. doi: 10.1016/j.virol.2009.03.016. Epub 2009 Apr 16. Virology. 2009. PMID: 19375143
-
The Lettuce infectious yellows virus (LIYV)-encoded P26 is associated with plasmalemma deposits within LIYV-infected cells.Virology. 2005 Mar 15;333(2):367-73. doi: 10.1016/j.virol.2005.01.012. Virology. 2005. PMID: 15721368
-
Cell-to-cell movement of plant viruses via plasmodesmata: a current perspective on potyviruses.Curr Opin Virol. 2021 Jun;48:10-16. doi: 10.1016/j.coviro.2021.03.002. Epub 2021 Mar 27. Curr Opin Virol. 2021. PMID: 33784579 Review.
-
Plant virus replication and movement.Virology. 2015 May;479-480:657-71. doi: 10.1016/j.virol.2015.01.025. Epub 2015 Mar 3. Virology. 2015. PMID: 25746797 Review.
Cited by
-
Killing two birds with one stone: How do Plant Viruses Break Down Plant Defenses and Manipulate Cellular Processes to Replicate Themselves?J Plant Biol. 2019;62(3):170-180. doi: 10.1007/s12374-019-0056-8. Epub 2019 May 27. J Plant Biol. 2019. PMID: 32218684 Free PMC article. Review.
-
Potyviral Helper-Component Protease: Multifaced Functions and Interactions with Host Proteins.Plants (Basel). 2024 Apr 29;13(9):1236. doi: 10.3390/plants13091236. Plants (Basel). 2024. PMID: 38732454 Free PMC article. Review.
-
Nonstructural p26 proteins encoded by the 3'-proximal genes of velariviruses and criniviruses are orthologs.Arch Virol. 2020 Feb;165(2):439-443. doi: 10.1007/s00705-019-04491-8. Epub 2019 Dec 11. Arch Virol. 2020. PMID: 31828509 Free PMC article.
-
Interactions of Tomato Chlorosis Virus p27 Protein with Tomato Catalase Are Involved in Viral Infection.Viruses. 2023 Apr 18;15(4):990. doi: 10.3390/v15040990. Viruses. 2023. PMID: 37112970 Free PMC article.
-
A functional investigation of the suppression of CpG and UpA dinucleotide frequencies in plant RNA virus genomes.Sci Rep. 2019 Dec 4;9(1):18359. doi: 10.1038/s41598-019-54853-0. Sci Rep. 2019. PMID: 31797900 Free PMC article.
References
-
- Agranovsky A. (2016). “Closteroviruses: molecular biology, evolution and interactions with cells,” in Plant Viruses: Evolution and Management, eds Gaur R. K., Petrov N. M., Patil B. L., Stoyanova M. I. (Singapore: Springer; ), 231–252. 10.1007/978-981-10-1406-2_14 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

