Polyprotein Processing as a Determinant for in Vitro Activity of Semliki Forest Virus Replicase
- PMID: 28991178
- PMCID: PMC5691643
- DOI: 10.3390/v9100292
Polyprotein Processing as a Determinant for in Vitro Activity of Semliki Forest Virus Replicase
Abstract
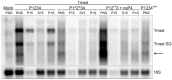
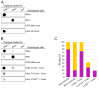
Semliki Forest virus (SFV) is an arthropod-borne alphavirus that induces membrane invaginations (spherules) in host cells. These harbor the viral replication complexes (RC) that synthesize viral RNA. Alphaviruses have four replicase or nonstructural proteins (nsPs), nsP1-4, expressed as polyprotein P1234. An early RC, which synthesizes minus-strand RNA, is formed by the polyprotein P123 and the polymerase nsP4. Further proteolytic cleavage results in a late RC consisting of nsP1-4 and synthesizing plus strands. Here, we show that only the late RCs are highly active in RNA synthesis in vitro. Furthermore, we demonstrate that active RCs can be isolated from both virus-infected cells and cells transfected with the wild-type replicase in combination with a plasmid expressing a template RNA. When an uncleavable polyprotein P123 and polymerase nsP4 were expressed together with a template, high levels of minus-strand RNA were produced in cells, but RCs isolated from these cells were hardly active in vitro. Furthermore, we observed that the uncleavable polyprotein P123 and polymerase nsP4, which have previously been shown to form spherules even in the absence of the template, did not replicate an exogenous template. Consequently, we hypothesize that the replicase proteins were sequestered in spherules and were no longer able to recruit a template.
Keywords: RNA synthesis; Semliki Forest virus; alphavirus; in vitro replication; nonstructural protein; polymerase; replication complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Regulation of Semliki Forest virus RNA replication: a model for the control of alphavirus pathogenesis in invertebrate hosts.Virology. 2004 May 20;323(1):153-63. doi: 10.1016/j.virol.2004.03.009. Virology. 2004. PMID: 15165827
-
Partially Uncleaved Alphavirus Replicase Forms Spherule Structures in the Presence and Absence of RNA Template.J Virol. 2017 Aug 24;91(18):e00787-17. doi: 10.1128/JVI.00787-17. Print 2017 Sep 15. J Virol. 2017. PMID: 28701392 Free PMC article.
-
Alphavirus polymerase and RNA replication.Virus Res. 2017 Apr 15;234:44-57. doi: 10.1016/j.virusres.2017.01.007. Epub 2017 Jan 16. Virus Res. 2017. PMID: 28104453 Review.
-
nsP4 Is a Major Determinant of Alphavirus Replicase Activity and Template Selectivity.J Virol. 2021 Sep 27;95(20):e0035521. doi: 10.1128/JVI.00355-21. Epub 2021 Jul 28. J Virol. 2021. PMID: 34319783 Free PMC article.
-
Alphavirus positive and negative strand RNA synthesis and the role of polyproteins in formation of viral replication complexes.Arch Virol Suppl. 1994;9:393-405. doi: 10.1007/978-3-7091-9326-6_39. Arch Virol Suppl. 1994. PMID: 8032270 Review.
Cited by
-
TMEM106A inhibits enveloped virus release from cell surface.iScience. 2022 Feb 1;25(2):103843. doi: 10.1016/j.isci.2022.103843. eCollection 2022 Feb 18. iScience. 2022. PMID: 35198896 Free PMC article.
-
TMEΜ45B Interacts with Sindbis Virus Nsp1 and Nsp4 and Inhibits Viral Replication.J Virol. 2022 Sep 14;96(17):e0091922. doi: 10.1128/jvi.00919-22. Epub 2022 Aug 8. J Virol. 2022. PMID: 35938871 Free PMC article.
-
A viral RNA motif involved in signaling the initiation of translation on non-AUG codons.RNA. 2019 Apr;25(4):431-452. doi: 10.1261/rna.068858.118. Epub 2019 Jan 18. RNA. 2019. PMID: 30659060 Free PMC article.
-
Purification of Highly Active Alphavirus Replication Complexes Demonstrates Altered Fractionation of Multiple Cellular Membranes.J Virol. 2018 Mar 28;92(8):e01852-17. doi: 10.1128/JVI.01852-17. Print 2018 Apr 15. J Virol. 2018. PMID: 29367248 Free PMC article.
-
Palmitoylated Cysteines in Chikungunya Virus nsP1 Are Critical for Targeting to Cholesterol-Rich Plasma Membrane Microdomains with Functional Consequences for Viral Genome Replication.J Virol. 2020 May 4;94(10):e02183-19. doi: 10.1128/JVI.02183-19. Print 2020 May 4. J Virol. 2020. PMID: 32132240 Free PMC article.
References
-
- King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J. Virus Taxonomy: Ninth Report of the International Commitee on Taxonmy of Viruses. Elsevier Academic Press; London, UK: 2012. pp. 10–14.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

