Cell cycle arrest in plants: what distinguishes quiescence, dormancy and differentiated G1?
- PMID: 28981580
- PMCID: PMC5737280
- DOI: 10.1093/aob/mcx082
Cell cycle arrest in plants: what distinguishes quiescence, dormancy and differentiated G1?
Abstract
Background: Quiescence is a fundamental feature of plant life, which enables plasticity, renewal and fidelity of the somatic cell line. Cellular quiescence is defined by arrest in a particular phase of the cell cycle, typically G1 or G2; however, the regulation of quiescence and proliferation can also be considered across wider scales in space and time. As such, quiescence is a defining feature of plant development and phenology, from meristematic stem cell progenitors to terminally differentiated cells, as well as dormant or suppressed seeds and buds. While the physiology of each of these states differs considerably, each is referred to as 'cell cycle arrest' or 'G1 arrest'.
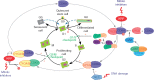
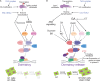
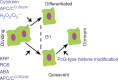
Scope: Here the physiology and molecular regulation of (1) meristematic quiescence, (2) dormancy and (3) terminal differentiation (cell cycle exit) are considered in order to determine whether and how the molecular decisions guiding these nuclear states are distinct. A brief overview of the canonical cell cycle regulators is provided, and the genetic and genomic, as well as physiological, evidence is considered regarding two primary questions: (1) Are the canonical cell cycle regulators superior or subordinate in the regulation of quiescence? (2) Are these three modes of quiescence governed by distinct molecular controls?
Conclusion: Meristematic quiescence, dormancy and terminal differentiation are each predominantly characterized by G1 arrest but regulated distinctly, at a level largely superior to the canonical cell cycle. Meristematic quiescence is intrinsically linked to non-cell-autonomous regulation of meristem cell identity, and particularly through the influence of ubiquitin-dependent proteolysis, in partnership with reactive oxygen species, abscisic acid and auxin. The regulation of terminal differentiation shares analogous features with meristematic quiescence, albeit with specific activators and a greater role for cytokinin signalling. Dormancy meanwhile appears to be regulated at the level of chromatin accessibility, by Polycomb group-type histone modifications of particular dormancy genes.
Keywords: Dormancy; branching; cell cycle; chromatin; differentiation; hormone; meristem; mitosis; proliferation; quiescence; reactive oxygen species; ubiquitin-dependent proteolysis.
© The Author 2017. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For Permissions, please email: journals.permissions@oup.com
Figures
Similar articles
-
Stem cell quiescence and dormancy in plant meristems.J Exp Bot. 2024 Oct 16;75(19):6022-6036. doi: 10.1093/jxb/erae201. J Exp Bot. 2024. PMID: 38721716 Free PMC article. Review.
-
On the language and physiology of dormancy and quiescence in plants.J Exp Bot. 2016 May;67(11):3189-203. doi: 10.1093/jxb/erw138. Epub 2016 Apr 6. J Exp Bot. 2016. PMID: 27053719 Review.
-
Cytokinin and the cell cycle.Curr Opin Plant Biol. 2014 Oct;21:7-15. doi: 10.1016/j.pbi.2014.05.015. Epub 2014 Jul 1. Curr Opin Plant Biol. 2014. PMID: 24994531 Review.
-
Temperature efficiency for dormancy release in apricot varies when applied at different amounts of chill accumulation.Plant Biol (Stuttg). 2013 Jan;15 Suppl 1:28-35. doi: 10.1111/j.1438-8677.2012.00636.x. Epub 2012 Jul 30. Plant Biol (Stuttg). 2013. PMID: 22845025
-
Developmental control of hypoxia during bud burst in grapevine.Plant Cell Environ. 2018 May;41(5):1154-1170. doi: 10.1111/pce.13141. Epub 2018 Feb 28. Plant Cell Environ. 2018. PMID: 29336037
Cited by
-
Multiple steps of leaf thickening during sun-leaf formation in Arabidopsis.Plant J. 2019 Nov;100(4):738-753. doi: 10.1111/tpj.14467. Epub 2019 Sep 9. Plant J. 2019. PMID: 31350790 Free PMC article.
-
Morphological and Ultrastructural Features of Formation of the Skin of Wheat (Triticum aestivum L.) Kernel.Plants (Basel). 2021 Nov 21;10(11):2538. doi: 10.3390/plants10112538. Plants (Basel). 2021. PMID: 34834901 Free PMC article.
-
Phytohormones: plant switchers in developmental and growth stages in potato.J Genet Eng Biotechnol. 2021 Jun 17;19(1):89. doi: 10.1186/s43141-021-00192-5. J Genet Eng Biotechnol. 2021. PMID: 34142228 Free PMC article. Review.
-
Beyond floral initiation: the role of flower bud dormancy in flowering time control of annual plants.J Exp Bot. 2024 Oct 16;75(19):6056-6062. doi: 10.1093/jxb/erae223. J Exp Bot. 2024. PMID: 38795335 Free PMC article. Review.
-
Metabolic regulation of quiescence in plants.Plant J. 2023 Jun;114(5):1132-1148. doi: 10.1111/tpj.16216. Epub 2023 Apr 13. Plant J. 2023. PMID: 36994639 Free PMC article.
References
-
- Aggarwal P, Padmanabhan B, Bhat A, Sarvepalli K, Sadhale PP, Nath U.. 2011. The TCP4 transcription factor of Arabidopsis blocks cell division in yeast at G1 → S transition. Biochemical and Biophysical Research Communications 410: 276–281. - PubMed
-
- Ahmed ZU, Kamra OP.. 1976. DNA content of dormant barley leaf nuclei and the rate of cell entry into S-phase and mitosis. Caryologia 29: 187–193.
-
- Aichinger E, Kornet N, Friedrich T, Laux T.. 2012. Plant stem cell niches. Annual Review of Plant Biology 63: 615–636. - PubMed
-
- Anh Tuan P, Bai S, Saito T, Imai T, Ito A, Moriguchi T.. 2016. Involvement of EARLY BUD-BREAK, an AP2/ERF transcription factor gene, in bud break in Japanese pear (Pyrus pyrifolia Nakai) lateral flower buds: expression, histone modifications and possible target genes. Plant and Cell Physiology 57: 1038–1047. - PubMed
-
- Armstrong W, Armstrong J.. 2014. Plant internal oxygen transport (diffusion and convection) and measuring and modelling oxygen gradients In: Van Dongen JT, Licausi F. eds. Low-oxygen stress in plants: oxygen sensing and adaptive responses to hypoxia. Vienna: Springer, 267–297.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

