Characterizing the structural ensemble of γ-secretase using a multiscale molecular dynamics approach
- PMID: 28970936
- PMCID: PMC5618787
- DOI: 10.1039/c7sc00980a
Characterizing the structural ensemble of γ-secretase using a multiscale molecular dynamics approach
Abstract
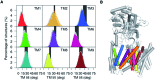
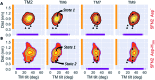
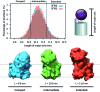
γ-Secretase is an intramembrane-cleaving aspartyl protease that plays an essential role in the processing of a variety of integral membrane proteins. Its role in the ultimate cleavage step in the processing of amyloid precursor protein to form amyloid-β (Aβ) peptide makes it an important therapeutic target in Alzheimer's disease research. Significant recent advances have been made in structural studies of this critical membrane protein complex. However, details of the mechanism of activation of the enzyme complex remain unclear. Using a multiscale computational modeling approach, combining multiple coarse-grained microsecond dynamic trajectories with all-atom models, the structure and two conformational states of the γ-secretase complex were evaluated. The transition between enzymatic state 1 and state 2 is shown to critically depend on the protonation states of the key catalytic residues Asp257 and Asp385 in the active site domain. The active site formation, related to our γ-secretase state 2, is observed to involve a concerted movement of four transmembrane helices from the catalytic subunit, resulting in the required localization of the catalytic residues. Global analysis of the structural ensemble of the enzyme complex was used to identify collective fluctuations important to the mechanism of substrate recognition and demonstrate that the corresponding fluctuations observed were uncorrelated with structural changes associated with enzyme activation. Overall, this computational study provides essential insight into the role of structure and dynamics in the activation and function of γ-secretase.
Figures
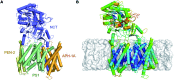


Similar articles
-
γ-Secretase Studied by Atomistic Molecular Dynamics Simulations: Global Dynamics, Enzyme Activation, Water Distribution and Lipid Binding.Front Chem. 2019 Jan 4;6:640. doi: 10.3389/fchem.2018.00640. eCollection 2018. Front Chem. 2019. PMID: 30662893 Free PMC article.
-
Active site geometry stabilization of a presenilin homolog by the lipid bilayer promotes intramembrane proteolysis.Elife. 2022 May 17;11:e76090. doi: 10.7554/eLife.76090. Elife. 2022. PMID: 35579427 Free PMC article.
-
Elucidating the Protonation State of the γ-Secretase Catalytic Dyad.ACS Chem Neurosci. 2023 Jan 18;14(2):261-269. doi: 10.1021/acschemneuro.2c00563. Epub 2022 Dec 23. ACS Chem Neurosci. 2023. PMID: 36562727
-
Inhibition of gamma-secretase as a therapeutic intervention for Alzheimer's disease: prospects, limitations and strategies.CNS Drugs. 2006;20(5):351-72. doi: 10.2165/00023210-200620050-00002. CNS Drugs. 2006. PMID: 16696577 Review.
-
Toward the structure of presenilin/γ-secretase and presenilin homologs.Biochim Biophys Acta. 2013 Dec;1828(12):2886-97. doi: 10.1016/j.bbamem.2013.04.015. Biochim Biophys Acta. 2013. PMID: 24099007 Free PMC article. Review.
Cited by
-
Structure and dynamics of γ-secretase with presenilin 2 compared to presenilin 1.RSC Adv. 2019 Jul 4;9(36):20901-20916. doi: 10.1039/c9ra02623a. eCollection 2019 Jul 1. RSC Adv. 2019. PMID: 35515530 Free PMC article.
-
Emerging structures and dynamic mechanisms of γ-secretase for Alzheimer's disease.Neural Regen Res. 2025 Jan 1;20(1):174-180. doi: 10.4103/NRR.NRR-D-23-01781. Epub 2024 Mar 1. Neural Regen Res. 2025. PMID: 38767485 Free PMC article.
-
Interaction of Substrates with γ-Secretase at the Level of Individual Transmembrane Helices-A Methodological Approach.Int J Mol Sci. 2023 Sep 21;24(18):14396. doi: 10.3390/ijms241814396. Int J Mol Sci. 2023. PMID: 37762696 Free PMC article.
-
Protein Predictive Modeling and Simulation of Mutations of Presenilin-1 Familial Alzheimer's Disease on the Orthosteric Site.Front Mol Biosci. 2021 Jun 2;8:649990. doi: 10.3389/fmolb.2021.649990. eCollection 2021. Front Mol Biosci. 2021. PMID: 34150846 Free PMC article.
-
Allosteric Modulation of Intact γ-Secretase Structural Dynamics.Biophys J. 2017 Dec 19;113(12):2634-2649. doi: 10.1016/j.bpj.2017.10.012. Biophys J. 2017. PMID: 29262358 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

