X chromosome inactivation in a female carrier of a 1.28 Mb deletion encompassing the human X inactivation centre
- PMID: 28947658
- PMCID: PMC5627161
- DOI: 10.1098/rstb.2016.0359
X chromosome inactivation in a female carrier of a 1.28 Mb deletion encompassing the human X inactivation centre
Abstract
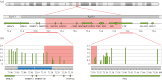
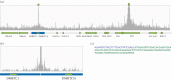
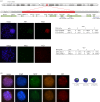
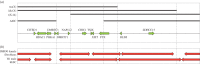
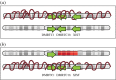
X chromosome inactivation (XCI) is a mechanism specifically initiated in female cells to silence one X chromosome, thereby equalizing the dose of X-linked gene products between male and female cells. XCI is regulated by a locus on the X chromosome termed the X-inactivation centre (XIC). Located within the XIC is XIST, which acts as a master regulator of XCI. During XCI, XIST is upregulated on the inactive X chromosome and chromosome-wide cis spreading of XIST leads to inactivation. In mouse, the Xic comprises Xist and all cis-regulatory elements and genes involved in Xist regulation. The activity of the XIC is regulated by trans-acting factors located elsewhere in the genome: X-encoded XCI activators positively regulating XCI, and autosomally encoded XCI inhibitors providing the threshold for XCI initiation. Whether human XCI is regulated through a similar mechanism, involving trans-regulatory factors acting on the XIC has remained elusive so far. Here, we describe a female individual with ovarian dysgenesis and a small X chromosomal deletion of the XIC. SNP-array and targeted locus amplification (TLA) analysis defined the deletion to a 1.28 megabase region, including XIST and all elements and genes that perform cis-regulatory functions in mouse XCI. Cells carrying this deletion still initiate XCI on the unaffected X chromosome, indicating that XCI can be initiated in the presence of only one XIC. Our results indicate that the trans-acting factors required for XCI initiation are located outside the deletion, providing evidence that the regulatory mechanisms of XCI are conserved between mouse and human.This article is part of the themed issue 'X-chromosome inactivation: a tribute to Mary Lyon'.
Keywords: X chromosome inactivation; XIC; XIST; deletion.
© 2017 The Authors.
Conflict of interest statement
We do not have any competing interests.
Figures

Similar articles
-
Loss of One X and the Y Chromosome Changes the Configuration of the X Inactivation Center in the Genus Tokudaia.Cytogenet Genome Res. 2024;164(1):23-32. doi: 10.1159/000539294. Epub 2024 May 16. Cytogenet Genome Res. 2024. PMID: 38754392
-
Xist and Tsix Transcription Dynamics Is Regulated by the X-to-Autosome Ratio and Semistable Transcriptional States.Mol Cell Biol. 2016 Oct 13;36(21):2656-2667. doi: 10.1128/MCB.00183-16. Print 2016 Nov 1. Mol Cell Biol. 2016. PMID: 27528619 Free PMC article.
-
The Ftx Noncoding Locus Controls X Chromosome Inactivation Independently of Its RNA Products.Mol Cell. 2018 May 3;70(3):462-472.e8. doi: 10.1016/j.molcel.2018.03.024. Mol Cell. 2018. PMID: 29706539
-
Regulation of X-chromosome inactivation in development in mice and humans.Microbiol Mol Biol Rev. 1998 Jun;62(2):362-78. doi: 10.1128/MMBR.62.2.362-378.1998. Microbiol Mol Biol Rev. 1998. PMID: 9618446 Free PMC article. Review.
-
Regulation of X-chromosome dosage compensation in human: mechanisms and model systems.Philos Trans R Soc Lond B Biol Sci. 2017 Nov 5;372(1733):20160363. doi: 10.1098/rstb.2016.0363. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28947660 Free PMC article. Review.
Cited by
-
Preface.Philos Trans R Soc Lond B Biol Sci. 2017 Nov 5;372(1733):20160353. doi: 10.1098/rstb.2016.0353. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28947653 Free PMC article. No abstract available.
References
-
- Lafreniere RG, Brown CJ, Rider S, Chelly J, Taillon-Miller P, Chinault AC, Monaco AP, Willard HF. 1993. 2.6 Mb YAC contig of the human X inactivation center region in Xq13: physical linkage of the RPS4X, PHKA1, XIST and DXS128E genes. Hum. Mol. Genet. 2, 1105–1115. (10.1093/hmg/2.8.1105) - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

