Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR
- PMID: 28935633
- PMCID: PMC5674890
- DOI: 10.1083/jcb.201703015
Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR
Abstract
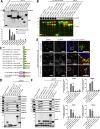
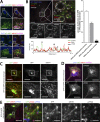
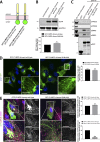
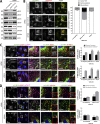
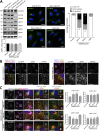
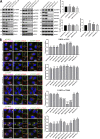
Endosomal recycling of transmembrane proteins requires sequence-dependent recognition of motifs present within their intracellular cytosolic domains. In this study, we have reexamined the role of retromer in the sequence-dependent endosome-to-trans-Golgi network (TGN) transport of the cation-independent mannose 6-phosphate receptor (CI-MPR). Although the knockdown or knockout of retromer does not perturb CI-MPR transport, the targeting of the retromer-linked sorting nexin (SNX)-Bin, Amphiphysin, and Rvs (BAR) proteins leads to a pronounced defect in CI-MPR endosome-to-TGN transport. The retromer-linked SNX-BAR proteins comprise heterodimeric combinations of SNX1 or SNX2 with SNX5 or SNX6 and serve to regulate the biogenesis of tubular endosomal sorting profiles. We establish that SNX5 and SNX6 associate with the CI-MPR through recognition of a specific WLM endosome-to-TGN sorting motif. From validating the CI-MPR dependency of SNX1/2-SNX5/6 tubular profile formation, we provide a mechanism for coupling sequence-dependent cargo recognition with the biogenesis of tubular profiles required for endosome-to-TGN transport. Therefore, the data presented in this study reappraise retromer's role in CI-MPR transport.
© 2017 Simonetti et al.
Figures



Comment in
-
Retromer revisited: Evolving roles for retromer in endosomal sorting.J Cell Biol. 2017 Nov 6;216(11):3433-3436. doi: 10.1083/jcb.201708111. Epub 2017 Oct 23. J Cell Biol. 2017. PMID: 29061649 Free PMC article.
Similar articles
-
Cargo-selective SNX-BAR proteins mediate retromer trimer independent retrograde transport.J Cell Biol. 2017 Nov 6;216(11):3677-3693. doi: 10.1083/jcb.201702137. Epub 2017 Sep 21. J Cell Biol. 2017. PMID: 28935632 Free PMC article.
-
Mechanism of cargo recognition by retromer-linked SNX-BAR proteins.PLoS Biol. 2020 Mar 9;18(3):e3000631. doi: 10.1371/journal.pbio.3000631. eCollection 2020 Mar. PLoS Biol. 2020. PMID: 32150533 Free PMC article.
-
Involvement of CASP9 (caspase 9) in IGF2R/CI-MPR endosomal transport.Autophagy. 2021 Jun;17(6):1393-1409. doi: 10.1080/15548627.2020.1761742. Epub 2020 May 25. Autophagy. 2021. PMID: 32397873 Free PMC article.
-
Retromer.Curr Opin Cell Biol. 2008 Aug;20(4):427-36. doi: 10.1016/j.ceb.2008.03.009. Epub 2008 May 9. Curr Opin Cell Biol. 2008. PMID: 18472259 Free PMC article. Review.
-
Retromer and sorting nexins in endosomal sorting.Biochem Soc Trans. 2015 Feb;43(1):33-47. doi: 10.1042/BST20140290. Biochem Soc Trans. 2015. PMID: 25619244 Review.
Cited by
-
TMEM16K is an interorganelle regulator of endosomal sorting.Nat Commun. 2020 Jul 3;11(1):3298. doi: 10.1038/s41467-020-17016-8. Nat Commun. 2020. PMID: 32620747 Free PMC article.
-
Toward Understanding the Molecular Role of SNX27/Retromer in Human Health and Disease.Front Cell Dev Biol. 2021 Apr 15;9:642378. doi: 10.3389/fcell.2021.642378. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33937239 Free PMC article. Review.
-
The retromer complex regulates C. elegans development and mammalian ciliogenesis.J Cell Sci. 2022 May 15;135(10):jcs259396. doi: 10.1242/jcs.259396. Epub 2022 May 17. J Cell Sci. 2022. PMID: 35510502 Free PMC article.
-
Retrograde transport of CDMPR depends on several machineries as analyzed by sulfatable nanobodies.Life Sci Alliance. 2022 Mar 21;5(7):e202101269. doi: 10.26508/lsa.202101269. Print 2022 Mar. Life Sci Alliance. 2022. PMID: 35314489 Free PMC article.
-
A novel live-cell imaging assay reveals regulation of endosome maturation.Elife. 2021 Nov 30;10:e70982. doi: 10.7554/eLife.70982. Elife. 2021. PMID: 34846303 Free PMC article.
References
-
- Carlton J., Bujny M., Peter B.J., Oorschot V.M., Rutherford A., Mellor H., Klumperman J., McMahon H.T., and Cullen P.J.. 2004. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr. Biol. 14:1791–1800. 10.1016/j.cub.2004.09.077 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

