Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites
- PMID: 28930685
- PMCID: PMC5646692
- DOI: 10.1016/j.celrep.2017.08.078
Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites
Abstract
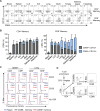
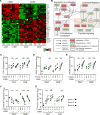
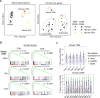
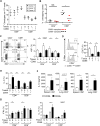
Tissue-resident memory T cells (TRMs) in mice mediate optimal protective immunity to infection and vaccination, while in humans, the existence and properties of TRMs remain unclear. Here, we use a unique human tissue resource to determine whether human tissue memory T cells constitute a distinct subset in diverse mucosal and lymphoid tissues. We identify a core transcriptional profile within the CD69+ subset of memory CD4+ and CD8+ T cells in lung and spleen that is distinct from that of CD69- TEM cells in tissues and circulation and defines human TRMs based on homology to the transcriptional profile of mouse CD8+ TRMs. Human TRMs in diverse sites exhibit increased expression of adhesion and inhibitory molecules, produce both pro-inflammatory and regulatory cytokines, and have reduced turnover compared with circulating TEM, suggesting unique adaptations for in situ immunity. Together, our results provide a unifying signature for human TRM and a blueprint for designing tissue-targeted immunotherapies.
Keywords: RNA-seq; human immunology; memory T cells; mucosal immunity.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Circulating memory CD8+ T cells are limited in forming CD103+ tissue-resident memory T cells at mucosal sites after reinfection.Eur J Immunol. 2021 Jan;51(1):151-166. doi: 10.1002/eji.202048737. Epub 2020 Aug 31. Eur J Immunol. 2021. PMID: 32762051
-
Functional heterogeneity of human tissue-resident memory T cells based on dye efflux capacities.JCI Insight. 2018 Nov 15;3(22):e123568. doi: 10.1172/jci.insight.123568. JCI Insight. 2018. PMID: 30429372 Free PMC article.
-
Cutting edge: resident memory CD8 T cells occupy frontline niches in secondary lymphoid organs.J Immunol. 2014 Apr 1;192(7):2961-4. doi: 10.4049/jimmunol.1400003. Epub 2014 Mar 5. J Immunol. 2014. PMID: 24600038 Free PMC article.
-
The Role of CD4+ Resident Memory T Cells in Local Immunity in the Mucosal Tissue - Protection Versus Pathology.Front Immunol. 2021 Apr 21;12:616309. doi: 10.3389/fimmu.2021.616309. eCollection 2021. Front Immunol. 2021. PMID: 33968018 Free PMC article. Review.
-
Functional Heterogeneity and Therapeutic Targeting of Tissue-Resident Memory T Cells.Cells. 2021 Jan 15;10(1):164. doi: 10.3390/cells10010164. Cells. 2021. PMID: 33467606 Free PMC article. Review.
Cited by
-
Toll-like receptor 7 protects against intestinal inflammation and restricts the development of colonic tissue-resident memory CD8+ T cells.Front Immunol. 2024 Oct 11;15:1465175. doi: 10.3389/fimmu.2024.1465175. eCollection 2024. Front Immunol. 2024. PMID: 39464882 Free PMC article.
-
Characterization of Nasal Mucosal T Cells in Horses and Their Response to Equine Herpesvirus Type 1.Viruses. 2024 Sep 25;16(10):1514. doi: 10.3390/v16101514. Viruses. 2024. PMID: 39459849 Free PMC article.
-
Evolutionary diversity of CXCL16-CXCR6: Convergent substitutions and recurrent gene loss in sauropsids.Immunogenetics. 2024 Dec;76(5-6):397-415. doi: 10.1007/s00251-024-01357-5. Epub 2024 Oct 14. Immunogenetics. 2024. PMID: 39400711
-
Injury-induced myosin-specific tissue-resident memory T cells drive immune checkpoint inhibitor myocarditis.Proc Natl Acad Sci U S A. 2024 Oct 15;121(42):e2323052121. doi: 10.1073/pnas.2323052121. Epub 2024 Oct 8. Proc Natl Acad Sci U S A. 2024. PMID: 39378095
-
Single-cell RNA sequencing reveals the pro-inflammatory roles of liver-resident Th1-like cells in primary biliary cholangitis.Nat Commun. 2024 Oct 7;15(1):8690. doi: 10.1038/s41467-024-53104-9. Nat Commun. 2024. PMID: 39375367 Free PMC article.
References
-
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

