Control of actin polymerization via the coincidence of phosphoinositides and high membrane curvature
- PMID: 28923975
- PMCID: PMC5674896
- DOI: 10.1083/jcb.201704061
Control of actin polymerization via the coincidence of phosphoinositides and high membrane curvature
Abstract
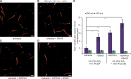
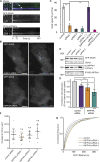
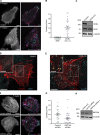
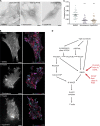
The conditional use of actin during clathrin-mediated endocytosis in mammalian cells suggests that the cell controls whether and how actin is used. Using a combination of biochemical reconstitution and mammalian cell culture, we elucidate a mechanism by which the coincidence of PI(4,5)P2 and PI(3)P in a curved vesicle triggers actin polymerization. At clathrin-coated pits, PI(3)P is produced by the INPP4A hydrolysis of PI(3,4)P2, and this is necessary for actin-driven endocytosis. Both Cdc42⋅guanosine triphosphate and SNX9 activate N-WASP-WIP- and Arp2/3-mediated actin nucleation. Membrane curvature, PI(4,5)P2, and PI(3)P signals are needed for SNX9 assembly via its PX-BAR domain, whereas signaling through Cdc42 is activated by PI(4,5)P2 alone. INPP4A activity is stimulated by high membrane curvature and synergizes with SNX9 BAR domain binding in a process we call curvature cascade amplification. We show that the SNX9-driven actin comets that arise on human disease-associated oculocerebrorenal syndrome of Lowe (OCRL) deficiencies are reduced by inhibiting PI(3)P production, suggesting PI(3)P kinase inhibitors as a therapeutic strategy in Lowe syndrome.
© 2017 Daste et al.
Figures
Similar articles
-
A role of OCRL in clathrin-coated pit dynamics and uncoating revealed by studies of Lowe syndrome cells.Elife. 2014 Aug 8;3:e02975. doi: 10.7554/eLife.02975. Elife. 2014. PMID: 25107275 Free PMC article.
-
Phosphoinositides and membrane curvature switch the mode of actin polymerization via selective recruitment of toca-1 and Snx9.Proc Natl Acad Sci U S A. 2013 Apr 30;110(18):7193-8. doi: 10.1073/pnas.1305286110. Epub 2013 Apr 15. Proc Natl Acad Sci U S A. 2013. PMID: 23589871 Free PMC article.
-
Triggering actin polymerization in Xenopus egg extracts from phosphoinositide-containing lipid bilayers.Methods Cell Biol. 2015;128:125-47. doi: 10.1016/bs.mcb.2015.01.020. Epub 2015 Apr 8. Methods Cell Biol. 2015. PMID: 25997346
-
Phosphoinositide regulation of clathrin-mediated endocytosis.Biochem Soc Trans. 2005 Dec;33(Pt 6):1285-9. doi: 10.1042/BST0331285. Biochem Soc Trans. 2005. PMID: 16246100 Review.
-
Phosphoinositides in endocytosis.Biochim Biophys Acta. 2015 Jun;1851(6):794-804. doi: 10.1016/j.bbalip.2014.09.014. Epub 2014 Sep 28. Biochim Biophys Acta. 2015. PMID: 25264171 Review.
Cited by
-
Modeling the neuropsychiatric manifestations of Lowe syndrome using induced pluripotent stem cells: defective F-actin polymerization and WAVE-1 expression in neuronal cells.Mol Autism. 2018 Aug 15;9:44. doi: 10.1186/s13229-018-0227-3. eCollection 2018. Mol Autism. 2018. PMID: 30147856 Free PMC article.
-
Phosphoinositide conversion in endocytosis and the endolysosomal system.J Biol Chem. 2018 Feb 2;293(5):1526-1535. doi: 10.1074/jbc.R117.000629. Epub 2017 Dec 27. J Biol Chem. 2018. PMID: 29282290 Free PMC article. Review.
-
PI(3,4)P2-mediated membrane tubulation promotes integrin trafficking and invasive cell migration.Proc Natl Acad Sci U S A. 2021 May 11;118(19):e2017645118. doi: 10.1073/pnas.2017645118. Proc Natl Acad Sci U S A. 2021. PMID: 33947811 Free PMC article.
-
Phosphatidylinositol 3,4-bisphosphate synthesis and turnover are spatially segregated in the endocytic pathway.J Biol Chem. 2020 Jan 24;295(4):1091-1104. doi: 10.1074/jbc.RA119.011774. Epub 2019 Dec 12. J Biol Chem. 2020. PMID: 31831620 Free PMC article.
-
Endosomal PI(3)P regulation by the COMMD/CCDC22/CCDC93 (CCC) complex controls membrane protein recycling.Nat Commun. 2019 Sep 19;10(1):4271. doi: 10.1038/s41467-019-12221-6. Nat Commun. 2019. PMID: 31537807 Free PMC article.
References
-
- Bago R., Malik N., Munson M.J., Prescott A.R., Davies P., Sommer E., Shpiro N., Ward R., Cross D., Ganley I.G., and Alessi D.R.. 2014. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J. 463:413–427. 10.1042/BJ20140889 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

