Omics Analyses of Trichoderma reesei CBS999.97 and QM6a Indicate the Relevance of Female Fertility to Carbohydrate-Active Enzyme and Transporter Levels
- PMID: 28916559
- PMCID: PMC5666144
- DOI: 10.1128/AEM.01578-17
Omics Analyses of Trichoderma reesei CBS999.97 and QM6a Indicate the Relevance of Female Fertility to Carbohydrate-Active Enzyme and Transporter Levels
Abstract
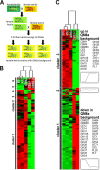
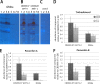
The filamentous fungus Trichoderma reesei is found predominantly in the tropics but also in more temperate regions, such as Europe, and is widely known as a producer of large amounts of plant cell wall-degrading enzymes. We sequenced the genome of the sexually competent isolate CBS999.97, which is phenotypically different from the female sterile strain QM6a but can cross sexually with QM6a. Transcriptome data for growth on cellulose showed that entire carbohydrate-active enzyme (CAZyme) families are consistently differentially regulated between these strains. We evaluated backcrossed strains of both mating types, which acquired female fertility from CBS999.97 but maintained a mostly QM6a genetic background, and we could thereby distinguish between the effects of strain background and female fertility or mating type. We found clear regulatory differences associated with female fertility and female sterility, including regulation of CAZyme and transporter genes. Analysis of carbon source utilization, transcriptomes, and secondary metabolites in these strains revealed that only a few changes in gene regulation are consistently correlated with different mating types. Different strain backgrounds (QM6a versus CBS999.97) resulted in the most significant alterations in the transcriptomes and in carbon source utilization, with decreased growth of CBS999.97 on several amino acids (for example proline or alanine), which further correlated with the downregulation of genes involved in the respective pathways. In combination, our findings support a role of fertility-associated processes in physiology and gene regulation and are of high relevance for the use of sexual crossing in combining the characteristics of two compatible strains or quantitative trait locus (QTL) analysis.IMPORTANCETrichoderma reesei is a filamentous fungus with a high potential for secretion of plant cell wall-degrading enzymes. We sequenced the genome of the fully fertile field isolate CBS999.97 and analyzed its gene regulation characteristics in comparison with the commonly used laboratory wild-type strain QM6a, which is not female fertile. Additionally, we also evaluated fully fertile strains with genotypes very close to that of QM6a in order to distinguish between strain-specific and fertility-specific characteristics. We found that QM6a and CBS999.97 clearly differ in their growth patterns on different carbon sources, CAZyme gene regulation, and secondary metabolism. Importantly, we found altered regulation of 90 genes associated with female fertility, including CAZyme genes and transporter genes, but only minor mating type-dependent differences. Hence, when using sexual crossing in research and for strain improvement, it is important to consider female fertile and female sterile strains for comparison with QM6a and to achieve optimal performance.
Keywords: Hypocrea jecorina; Trichoderma reesei; carbon source; cellulase; female fertility; mating type; secondary metabolism; sexual development; strain improvement.
Copyright © 2017 Tisch et al.
Figures
Similar articles
-
Trichoderma reesei Isolated From Austrian Soil With High Potential for Biotechnological Application.Front Microbiol. 2021 Jan 28;12:552301. doi: 10.3389/fmicb.2021.552301. eCollection 2021. Front Microbiol. 2021. PMID: 33584603 Free PMC article.
-
Analysis of Light- and Carbon-Specific Transcriptomes Implicates a Class of G-Protein-Coupled Receptors in Cellulose Sensing.mSphere. 2017 May 10;2(3):e00089-17. doi: 10.1128/mSphere.00089-17. eCollection 2017 May-Jun. mSphere. 2017. PMID: 28497120 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
Cited by
-
The G-protein Coupled Receptor GPR8 Regulates Secondary Metabolism in Trichoderma reesei.Front Bioeng Biotechnol. 2020 Nov 5;8:558996. doi: 10.3389/fbioe.2020.558996. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33251193 Free PMC article.
-
Gene regulation associated with sexual development and female fertility in different isolates of Trichoderma reesei.Fungal Biol Biotechnol. 2018 May 15;5:9. doi: 10.1186/s40694-018-0055-4. eCollection 2018. Fungal Biol Biotechnol. 2018. PMID: 29785273 Free PMC article.
-
Trichoderma reesei Isolated From Austrian Soil With High Potential for Biotechnological Application.Front Microbiol. 2021 Jan 28;12:552301. doi: 10.3389/fmicb.2021.552301. eCollection 2021. Front Microbiol. 2021. PMID: 33584603 Free PMC article.
-
Transgressive phenotypes from outbreeding between the Trichoderma reesei hyper producer RutC30 and a natural isolate.Microbiol Spectr. 2024 Oct 3;12(10):e0044124. doi: 10.1128/spectrum.00441-24. Epub 2024 Aug 20. Microbiol Spectr. 2024. PMID: 39162516 Free PMC article.
-
Protein hyperproduction in fungi by design.Appl Microbiol Biotechnol. 2018 Oct;102(20):8621-8628. doi: 10.1007/s00253-018-9265-1. Epub 2018 Aug 4. Appl Microbiol Biotechnol. 2018. PMID: 30078136 Free PMC article. Review.
References
-
- Schmoll M, Seiboth B, Druzhinina I, Kubicek CP. 2014. Genomics analysis of biocontrol species and industrial enzyme producers from the genus Trichoderma, p 233–266. In Esser K, Nowrousian M (ed), The Mycota XIII. Springer, Berlin, Germany.
-
- Paloheimo M, Haarmann T, Mäkinen S, Vehmaanperä J. 2016. Production of industrial enzymes in Trichoderma reesei, p 23–58. In Schmoll M, Dattenb̈ock C (ed), Gene expression systems in fungi: advancements and applications. Springer International, Heidelberg, Germany.
-
- Schmoll M, Dattenbock C, Carreras-Villasenor N, Mendoza-Mendoza A, Tisch D, Aleman MI, Baker SE, Brown C, Cervantes-Badillo MG, Cetz-Chel J, Cristobal-Mondragon GR, Delaye L, Esquivel-Naranjo EU, Frischmann A, Gallardo-Negrete Jde J, Garcia-Esquivel M, Gomez-Rodriguez EY, Greenwood DR, Hernandez-Onate M, Kruszewska JS, Lawry R, Mora-Montes HM, Munoz-Centeno T, Nieto-Jacobo MF, Nogueira Lopez G, Olmedo-Monfil V, Osorio-Concepcion M, Pilsyk S, Pomraning KR, Rodriguez-Iglesias A, Rosales-Saavedra MT, Sanchez-Arreguin JA, Seidl-Seiboth V, Stewart A, Uresti-Rivera EE, Wang CL, Wang TF, Zeilinger S, Casas-Flores S, Herrera-Estrella A. 2016. The genomes of three uneven siblings: footprints of the lifestyles of three Trichoderma species. Microbiol Mol Biol Rev 80:205–327. doi:10.1128/MMBR.00040-15. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

