Non-polio enteroviruses in faeces of children diagnosed with acute flaccid paralysis in Nigeria
- PMID: 28899411
- PMCID: PMC5596853
- DOI: 10.1186/s12985-017-0846-x
Non-polio enteroviruses in faeces of children diagnosed with acute flaccid paralysis in Nigeria
Abstract
Background: The need to investigate the contribution of non-polio enteroviruses to acute flaccid paralysis (AFP) cannot be over emphasized as we move towards a poliovirus free world. Hence, we aim to identify non-polio enteroviruses recovered from the faeces of children diagnosed with AFP in Nigeria.
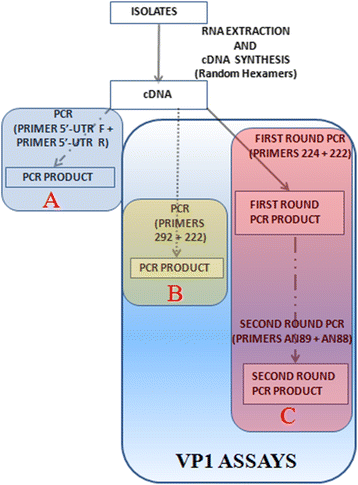
Methods: Ninety-six isolates, (95 unidentified and one previously confirmed Sabin poliovirus 3) recovered on RD cell culture from the stool of children <15 years old diagnosed with AFP in 2014 were analyzed. All isolates were subjected to RNA extraction, cDNA synthesis and three different PCR reactions (one panenterovirus 5'-UTR and two different VP1 amplification assays). VP1 amplicons were then sequenced and isolates identified.
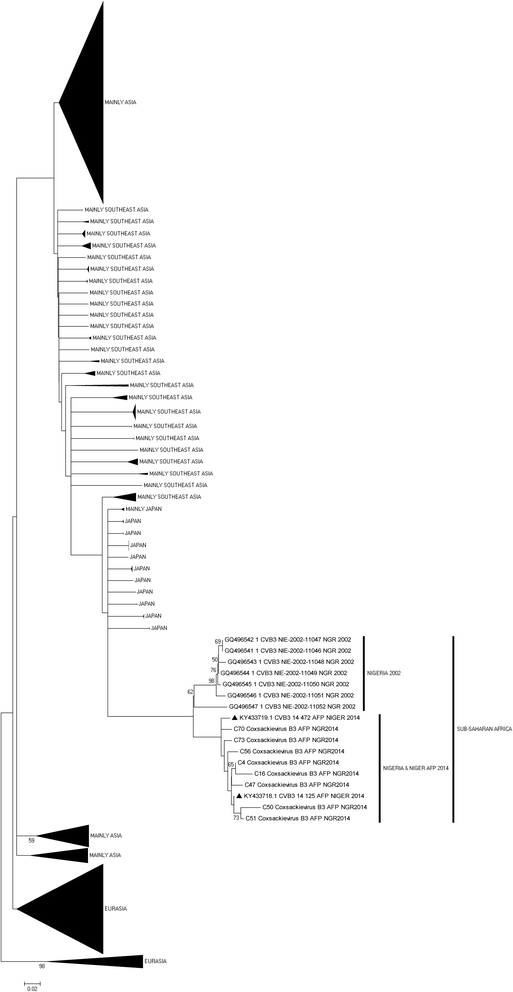
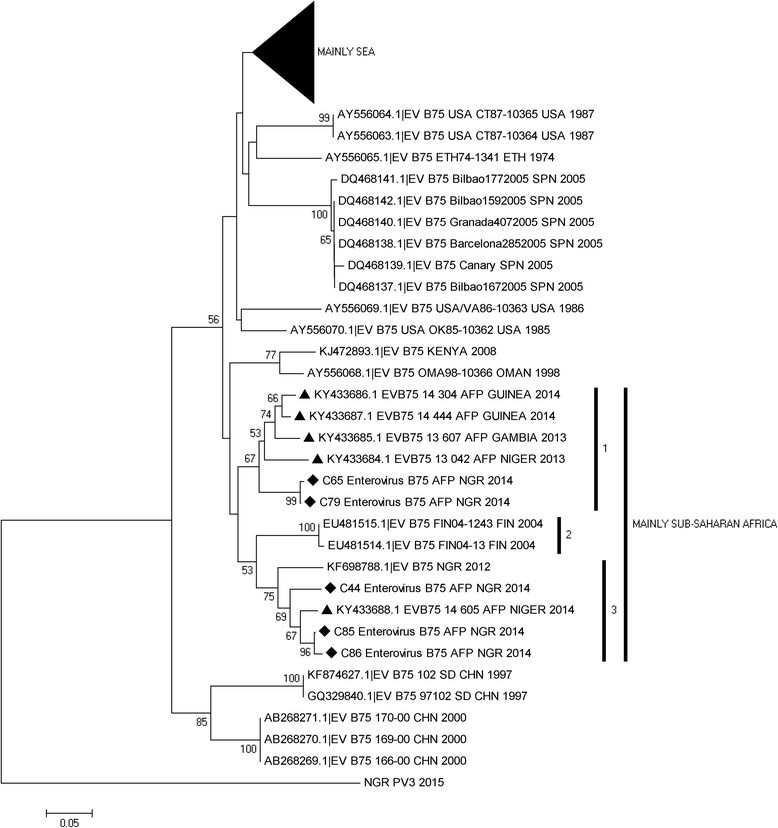
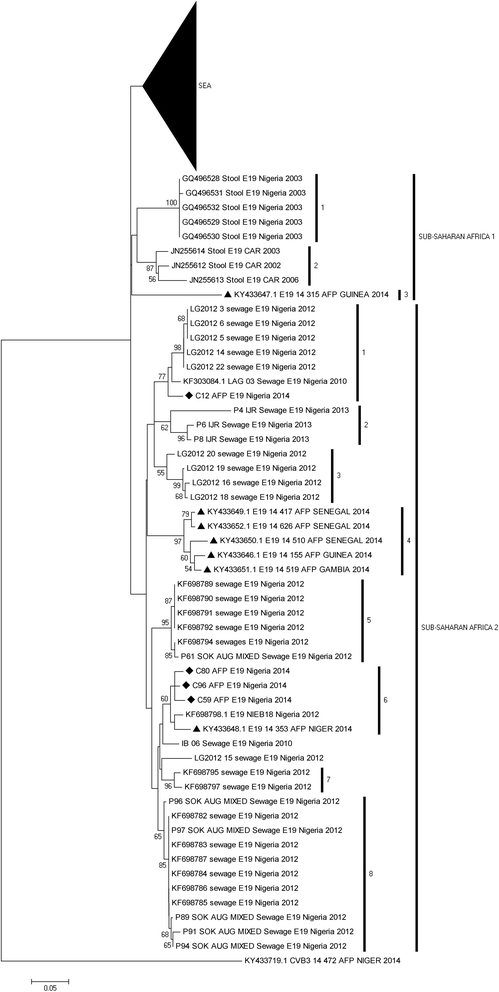
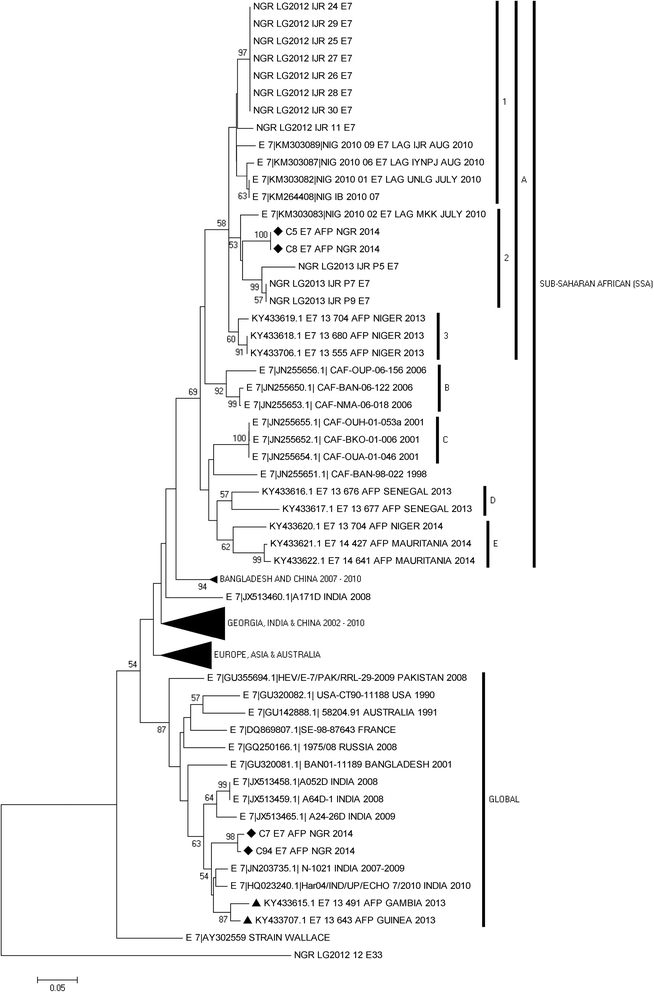
Results: 92.71% (89/96) of the isolates were detected by at least one of the three assays as an enterovirus. Precisely, 79.17% (76/96), 6.25% (6/96), 7.30% (7/96) and 7.30% (7/96) of the isolates were positive for both, positive and negative, negative and positive, as well as negative for both the 5'-UTR and VP1 assays, respectively. In this study, sixty-nine (69) of the 83 VP1 amplicons sequenced were identified as 27 different enterovirus types. The most commonly detected were CV-B3 (10 isolates) and EV-B75 (5 isolates). Specifically, one, twenty-four and two of the enterovirus types identified in this study belong to EV-A, EV-B and EV-C respectively.
Conclusions: This study reports the circulating strains of 27 non-polio enterovirus types in Nigerian children with AFP in 2014 and Nigerian strains of CV-B2, CV-B4, E17, EV-B80, EV-B73, EV-B97, EV-B93, EV-C99 and EV-A120 were reported for the first time. Furthermore, it shows that being positive for the 5'-UTR assay should not be the basis for subjecting isolates to the VP1 assays.
Keywords: AFP; Enteroviruses; Nigeria; Non-polio enteroviruses; VP1 analysis.
Conflict of interest statement
Ethics approval and consent to participate
Enterovirus isolates were analysed in this study. The stool samples from which these isolates were recovered were collected in accordance with the national ethical guidelines as part of the National AFP surveillance programme in Nigeria and sent to the WHO National Polio Laboratory in Ibadan, Nigeria to ascertain whether poliovirus is the etiologic agent of the diagnosed AFP using the WHO algorithm. The isolates analyzed in this study were subsequently anonymized for further studies before use in this study. Thus, this article does not contain any studies with human participants performed by any of the authors. In addition, no information that can be used to associate the isolates analyzed in this study to any individual is included in this manuscript.
Consent for publication
Not applicable.
Competing interests
The authors declare that no conflict of interests exist.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Identification of previously untypable RD cell line isolates and detection of EV-A71 genotype C1 in a child with AFP in Nigeria.Pathog Glob Health. 2018 Dec;112(8):421-427. doi: 10.1080/20477724.2018.1548117. Epub 2018 Nov 26. Pathog Glob Health. 2018. PMID: 30474520 Free PMC article.
-
New enteroviruses, EV-93 and EV-94, associated with acute flaccid paralysis in the Democratic Republic of the Congo.J Med Virol. 2007 Apr;79(4):393-400. doi: 10.1002/jmv.20825. J Med Virol. 2007. PMID: 17311342
-
Preponderance of enterovirus C in RD-L20B-cell-culture-negative stool samples from children diagnosed with acute flaccid paralysis in Nigeria.Arch Virol. 2017 Oct;162(10):3089-3101. doi: 10.1007/s00705-017-3466-2. Epub 2017 Jul 10. Arch Virol. 2017. PMID: 28691129
-
Non-polio Enterovirus detection with acute flaccid paralysis: A systematic review.J Med Virol. 2018 Jan;90(1):3-7. doi: 10.1002/jmv.24933. Epub 2017 Oct 12. J Med Virol. 2018. PMID: 28857219 Review.
-
Recommendations for enterovirus diagnostics and characterisation within and beyond Europe.J Clin Virol. 2018 Apr;101:11-17. doi: 10.1016/j.jcv.2018.01.008. Epub 2018 Feb 6. J Clin Virol. 2018. PMID: 29414181 Review.
Cited by
-
Antiviral potentials of Lactobacillus plantarum, Lactobacillus amylovorus, and Enterococcus hirae against selected Enterovirus.Folia Microbiol (Praha). 2019 Mar;64(2):257-264. doi: 10.1007/s12223-018-0648-6. Epub 2018 Sep 29. Folia Microbiol (Praha). 2019. PMID: 30267215
-
Molecular typing and characterization of a novel genotype of EV-B93 isolated from Tibet, China.PLoS One. 2020 Aug 25;15(8):e0237652. doi: 10.1371/journal.pone.0237652. eCollection 2020. PLoS One. 2020. PMID: 32841272 Free PMC article.
-
High Diversity of Human Non-Polio Enterovirus Serotypes Identified in Contaminated Water in Nigeria.Viruses. 2021 Feb 5;13(2):249. doi: 10.3390/v13020249. Viruses. 2021. PMID: 33562806 Free PMC article.
-
Molecular characterization and epidemiological aspects of non-polio enteroviruses isolated from acute flaccid paralysis in Brazil: a historical series (2005-2017).Emerg Microbes Infect. 2020 Dec;9(1):2536-2546. doi: 10.1080/22221751.2020.1850181. Emerg Microbes Infect. 2020. PMID: 33179584 Free PMC article.
-
Metagenomic surveillance uncovers diverse and novel viral taxa in febrile patients from Nigeria.Nat Commun. 2023 Aug 4;14(1):4693. doi: 10.1038/s41467-023-40247-4. Nat Commun. 2023. PMID: 37542071 Free PMC article.
References
-
- Oberste MS, Maher K, Williams AJ, Dybdahl-Sissoko N, Brown BA, Gookin MS, Penaranda S, Mishrik N, Uddin M, Pallansch MA. Species-specific RT-PCR amplification of human enteroviruses: a tool for rapid species identification of uncharacterized enteroviruses. J Gen Virol. 2006;87:119–128. doi: 10.1099/vir.0.81179-0. - DOI - PubMed
-
- Oberste MS, Pallansch MA. Enteroviruses molecular detection and typing. Reviews in Medical Microbiology. 2005;16:163–171. doi: 10.1097/01.revmedmi.0000184741.90926.35. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

