Global Analysis of Membrane-associated Protein Oligomerization Using Protein Correlation Profiling
- PMID: 28887381
- PMCID: PMC5672003
- DOI: 10.1074/mcp.RA117.000276
Global Analysis of Membrane-associated Protein Oligomerization Using Protein Correlation Profiling
Abstract
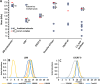
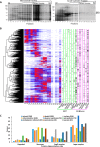
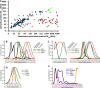
Membrane-associated proteins are required for essential processes like transport, organelle biogenesis, and signaling. Many are expected to function as part of an oligomeric protein complex. However, membrane-associated proteins are challenging to work with, and large-scale data sets on the oligomerization state of this important class of proteins is missing. Here we combined cell fractionation of Arabidopsis leaves with nondenaturing detergent solubilization and LC/MS-based profiling of size exclusion chromatography fractions to measure the apparent masses of >1350 membrane-associated proteins. Our method identified proteins from all of the major organelles, with more than 50% of them predicted to be part of a stable complex. The plasma membrane was the most highly enriched in large protein complexes compared with other organelles. Hundreds of novel protein complexes were identified. Over 150 proteins had a complicated localization pattern, and were clearly partitioned between cytosolic and membrane-associated pools. A subset of these dual localized proteins had oligomerization states that differed based on localization. Our data set is an important resource for the community that includes new functionally relevant data for membrane-localized protein complexes that could not be predicted based on sequence alone. Our method enables the analysis of protein complex localization and dynamics, and is a first step in the development of a method in which LC/MS profile data can be used to predict the composition of membrane-associated protein complexes.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

Similar articles
-
Analysis of protein complexes in Arabidopsis leaves using size exclusion chromatography and label-free protein correlation profiling.J Proteomics. 2017 Aug 23;166:8-18. doi: 10.1016/j.jprot.2017.06.004. Epub 2017 Jun 13. J Proteomics. 2017. PMID: 28627464
-
A proteomic strategy for global analysis of plant protein complexes.Plant Cell. 2014 Oct;26(10):3867-82. doi: 10.1105/tpc.114.127563. Epub 2014 Oct 7. Plant Cell. 2014. PMID: 25293756 Free PMC article.
-
A Label-free Mass Spectrometry Method to Predict Endogenous Protein Complex Composition.Mol Cell Proteomics. 2019 Aug;18(8):1588-1606. doi: 10.1074/mcp.RA119.001400. Epub 2019 Jun 11. Mol Cell Proteomics. 2019. PMID: 31186290 Free PMC article.
-
Specific interaction of IM30/Vipp1 with cyanobacterial and chloroplast membranes results in membrane remodeling and eventually in membrane fusion.Biochim Biophys Acta Biomembr. 2017 Apr;1859(4):537-549. doi: 10.1016/j.bbamem.2016.09.025. Epub 2016 Sep 30. Biochim Biophys Acta Biomembr. 2017. PMID: 27693914 Review.
-
The proteomics of plant cell membranes.J Exp Bot. 2007;58(1):103-12. doi: 10.1093/jxb/erj209. Epub 2006 Jun 27. J Exp Bot. 2007. PMID: 16804056 Review.
Cited by
-
Connecting the dots: from nanodomains to physiological functions of REMORINs.Plant Physiol. 2021 Apr 2;185(3):632-649. doi: 10.1093/plphys/kiaa063. Plant Physiol. 2021. PMID: 33793872 Free PMC article. Review.
-
Interactome determination of a Long Noncoding RNA implicated in Embryonic Stem Cell Self-Renewal.Sci Rep. 2018 Dec 4;8(1):17568. doi: 10.1038/s41598-018-34864-z. Sci Rep. 2018. PMID: 30514857 Free PMC article.
-
Complexome Profiling Reveals Association of PPR Proteins with Ribosomes in the Mitochondria of Plants.Mol Cell Proteomics. 2019 Jul;18(7):1345-1362. doi: 10.1074/mcp.RA119.001396. Epub 2019 Apr 25. Mol Cell Proteomics. 2019. PMID: 31023727 Free PMC article.
-
FLA11 and FLA12 glycoproteins fine-tune stem secondary wall properties in response to mechanical stresses.New Phytol. 2022 Feb;233(4):1750-1767. doi: 10.1111/nph.17898. Epub 2022 Jan 4. New Phytol. 2022. PMID: 34862967 Free PMC article.
-
Tandem Mass Tagging-Based Quantitative Proteomics Analysis Reveals Damage to the Liver and Brain of Hypophthalmichthys molitrix Exposed to Acute Hypoxia and Reoxygenation.Antioxidants (Basel). 2022 Mar 19;11(3):589. doi: 10.3390/antiox11030589. Antioxidants (Basel). 2022. PMID: 35326239 Free PMC article.
References
-
- Kush G. S. (2001) Green revolution: the way forward. Nat. Rev. Genet. 2, 815–822 - PubMed
-
- Hartwell L., Hopfield J., Leibler S., and Murray A. (1999) From molecular to modular cell biology. Nature 402, C47–C52 - PubMed
-
- Alberts B. (1998) The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92, 291–294 - PubMed
-
- Berg J. M., Tymoczko J. L., Gatto G. J. Jr, and Stryer L. (2015) Biochemistry (W. H. Freeman, New York: ). 8th Ed.
-
- Buchanan B. B., Gruissem W., and Jones R. L. (2015) Biochemistry and Molecular Biology of Plants (Whiley) 2nd Ed.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

