Revisiting the Role of Csp Family Proteins in Regulating Clostridium difficile Spore Germination
- PMID: 28874406
- PMCID: PMC5648855
- DOI: 10.1128/JB.00266-17
Revisiting the Role of Csp Family Proteins in Regulating Clostridium difficile Spore Germination
Abstract
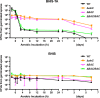
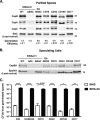
Clostridium difficile causes considerable health care-associated gastrointestinal disease that is transmitted by its metabolically dormant spore form. Upon entering the gut, C. difficile spores germinate and outgrow to produce vegetative cells that release disease-causing toxins. C. difficile spore germination depends on the Csp family of (pseudo)proteases and the cortex hydrolase SleC. The CspC pseudoprotease functions as a bile salt germinant receptor that activates the protease CspB, which in turn proteolytically activates the SleC zymogen. Active SleC degrades the protective cortex layer, allowing spores to outgrow and resume metabolism. We previously showed that the CspA pseudoprotease domain, which is initially produced as a fusion to CspB, controls the incorporation of the CspC germinant receptor in mature spores. However, study of the individual Csp proteins has been complicated by the polar effects of TargeTron-based gene disruption on the cspBA-cspC operon. To overcome these limitations, we have used pyrE-based allelic exchange to create individual deletions of the regions encoding CspB, CspA, CspBA, and CspC in strain 630Δerm Our results indicate that stable CspA levels in sporulating cells depend on CspB and confirm that CspA maximizes CspC incorporation into spores. Interestingly, we observed that csp and sleC mutants spontaneously germinate more frequently in 630Δerm than equivalent mutants in the JIR8094 and UK1 strain backgrounds. Analyses of this phenomenon suggest that only a subpopulation of C. difficile 630Δerm spores can spontaneously germinate, in contrast with Bacillus subtilis spores. We also show that C. difficile clinical isolates that encode truncated CspBA variants have sequencing errors that actually produce full-length CspBA.IMPORTANCEClostridium difficile is a leading cause of health care-associated infections. Initiation of C. difficile infection depends on spore germination, a process controlled by Csp family (pseudo)proteases. The CspC pseudoprotease is a germinant receptor that senses bile salts and activates the CspB protease, which activates a hydrolase required for germination. Previous work implicated the CspA pseudoprotease in controlling CspC incorporation into spores but relied on plasmid-based overexpression. Here we have used allelic exchange to study the functions of CspB and CspA. We determined that CspA production and/or stability depends on CspB and confirmed that CspA maximizes CspC incorporation into spores. Our data also suggest that a subpopulation of C. difficile spores spontaneously germinates in the absence of bile salt germinants and/or Csp proteins.
Keywords: Clostridium difficile; Csp; germination; spontaneous germination; spores.
Copyright © 2017 American Society for Microbiology.
Figures


Similar articles
-
Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain.Biochimie. 2016 Mar;122:243-54. doi: 10.1016/j.biochi.2015.07.023. Epub 2015 Jul 29. Biochimie. 2016. PMID: 26231446 Free PMC article.
-
A Clostridium difficile-Specific, Gel-Forming Protein Required for Optimal Spore Germination.mBio. 2017 Jan 17;8(1):e02085-16. doi: 10.1128/mBio.02085-16. mBio. 2017. PMID: 28096487 Free PMC article.
-
The requirement for co-germinants during Clostridium difficile spore germination is influenced by mutations in yabG and cspA.PLoS Pathog. 2019 Apr 3;15(4):e1007681. doi: 10.1371/journal.ppat.1007681. eCollection 2019 Apr. PLoS Pathog. 2019. PMID: 30943268 Free PMC article.
-
A Revised Understanding of Clostridioides difficile Spore Germination.Trends Microbiol. 2020 Sep;28(9):744-752. doi: 10.1016/j.tim.2020.03.004. Epub 2020 Apr 23. Trends Microbiol. 2020. PMID: 32781028 Review.
-
Clostridium difficile spore germination: an update.Res Microbiol. 2010 Nov;161(9):730-4. doi: 10.1016/j.resmic.2010.09.007. Epub 2010 Sep 21. Res Microbiol. 2010. PMID: 20863888 Review.
Cited by
-
Mechanism of germination inhibition of Clostridioides difficile spores by an aniline substituted cholate derivative (CaPA).J Antibiot (Tokyo). 2023 Jun;76(6):335-345. doi: 10.1038/s41429-023-00612-3. Epub 2023 Apr 4. J Antibiot (Tokyo). 2023. PMID: 37016015 Free PMC article.
-
The impact of YabG mutations on C. difficile spore germination and processing of spore substrates.bioRxiv [Preprint]. 2024 Jun 10:2024.06.10.598338. doi: 10.1101/2024.06.10.598338. bioRxiv. 2024. Update in: Mol Microbiol. 2024 Oct;122(4):534-548. doi: 10.1111/mmi.15316. PMID: 38915615 Free PMC article. Updated. Preprint.
-
Complete Genome Sequencing and Comparative Phenotypic Analysis Reveal the Discrepancy Between Clostridioides difficile ST81 and ST37 Isolates.Front Microbiol. 2021 Dec 21;12:776892. doi: 10.3389/fmicb.2021.776892. eCollection 2021. Front Microbiol. 2021. PMID: 34992586 Free PMC article.
-
Studies on the Importance of the 7α-, and 12α- hydroxyl groups of N-Aryl-3α,7α,12α-trihydroxy-5β-cholan-24-amides on their Antigermination Activity Against a Hypervirulent Strain of Clostridioides (Clostridium) difficile.Bioorg Med Chem. 2021 Dec 15;52:116503. doi: 10.1016/j.bmc.2021.116503. Epub 2021 Nov 10. Bioorg Med Chem. 2021. PMID: 34837818 Free PMC article.
-
Sporulation and Germination in Clostridial Pathogens.Microbiol Spectr. 2019 Nov;7(6):10.1128/microbiolspec.gpp3-0017-2018. doi: 10.1128/microbiolspec.GPP3-0017-2018. Microbiol Spectr. 2019. PMID: 31858953 Free PMC article. Review.
References
-
- Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/threat-report-2013.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

