5C analysis of the Epidermal Differentiation Complex locus reveals distinct chromatin interaction networks between gene-rich and gene-poor TADs in skin epithelial cells
- PMID: 28863138
- PMCID: PMC5599062
- DOI: 10.1371/journal.pgen.1006966
5C analysis of the Epidermal Differentiation Complex locus reveals distinct chromatin interaction networks between gene-rich and gene-poor TADs in skin epithelial cells
Abstract
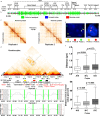
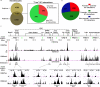
Mammalian genomes contain several dozens of large (>0.5 Mbp) lineage-specific gene loci harbouring functionally related genes. However, spatial chromatin folding, organization of the enhancer-promoter networks and their relevance to Topologically Associating Domains (TADs) in these loci remain poorly understood. TADs are principle units of the genome folding and represents the DNA regions within which DNA interacts more frequently and less frequently across the TAD boundary. Here, we used Chromatin Conformation Capture Carbon Copy (5C) technology to characterize spatial chromatin interaction network in the 3.1 Mb Epidermal Differentiation Complex (EDC) locus harbouring 61 functionally related genes that show lineage-specific activation during terminal keratinocyte differentiation in the epidermis. 5C data validated by 3D-FISH demonstrate that the EDC locus is organized into several TADs showing distinct lineage-specific chromatin interaction networks based on their transcription activity and the gene-rich or gene-poor status. Correlation of the 5C results with genome-wide studies for enhancer-specific histone modifications (H3K4me1 and H3K27ac) revealed that the majority of spatial chromatin interactions that involves the gene-rich TADs at the EDC locus in keratinocytes include both intra- and inter-TAD interaction networks, connecting gene promoters and enhancers. Compared to thymocytes in which the EDC locus is mostly transcriptionally inactive, these interactions were found to be keratinocyte-specific. In keratinocytes, the promoter-enhancer anchoring regions in the gene-rich transcriptionally active TADs are enriched for the binding of chromatin architectural proteins CTCF, Rad21 and chromatin remodeler Brg1. In contrast to gene-rich TADs, gene-poor TADs show preferential spatial contacts with each other, do not contain active enhancers and show decreased binding of CTCF, Rad21 and Brg1 in keratinocytes. Thus, spatial interactions between gene promoters and enhancers at the multi-TAD EDC locus in skin epithelial cells are cell type-specific and involve extensive contacts within TADs as well as between different gene-rich TADs, forming the framework for lineage-specific transcription.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells.Development. 2014 Jan;141(1):101-11. doi: 10.1242/dev.103200. Development. 2014. PMID: 24346698 Free PMC article.
-
Multiple CTCF sites cooperate with each other to maintain a TAD for enhancer-promoter interaction in the β-globin locus.FASEB J. 2021 Aug;35(8):e21768. doi: 10.1096/fj.202100105RR. FASEB J. 2021. PMID: 34245617
-
Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding.Mol Cell. 2020 May 7;78(3):539-553.e8. doi: 10.1016/j.molcel.2020.03.002. Epub 2020 Mar 25. Mol Cell. 2020. PMID: 32213323 Free PMC article.
-
The Interplay Between Chromatin Architecture and Lineage-Specific Transcription Factors and the Regulation of Rag Gene Expression.Front Immunol. 2021 Mar 16;12:659761. doi: 10.3389/fimmu.2021.659761. eCollection 2021. Front Immunol. 2021. PMID: 33796120 Free PMC article. Review.
-
Pushing the TAD boundary: Decoding insulator codes of clustered CTCF sites in 3D genomes.Bioessays. 2024 Oct;46(10):e2400121. doi: 10.1002/bies.202400121. Epub 2024 Aug 21. Bioessays. 2024. PMID: 39169755 Review.
Cited by
-
A Kaleidoscope of Keratin Gene Expression and the Mosaic of Its Regulatory Mechanisms.Int J Mol Sci. 2023 Mar 15;24(6):5603. doi: 10.3390/ijms24065603. Int J Mol Sci. 2023. PMID: 36982676 Free PMC article. Review.
-
Overview of the molecular determinants contributing to the expression of Psoriasis and Psoriatic Arthritis phenotypes.J Cell Mol Med. 2020 Dec;24(23):13554-13563. doi: 10.1111/jcmm.15742. Epub 2020 Oct 31. J Cell Mol Med. 2020. PMID: 33128843 Free PMC article. Review.
-
Chromatin Landscape Governing Murine Epidermal Differentiation.J Invest Dermatol. 2023 Jul;143(7):1220-1232.e9. doi: 10.1016/j.jid.2022.12.020. Epub 2023 Jan 26. J Invest Dermatol. 2023. PMID: 36708949 Free PMC article.
-
Cohesin mediates Esco2-dependent transcriptional regulation in a zebrafish regenerating fin model of Roberts Syndrome.Biol Open. 2017 Dec 15;6(12):1802-1813. doi: 10.1242/bio.026013. Biol Open. 2017. PMID: 29084713 Free PMC article.
-
Tales from topographic oceans: topologically associated domains and cancer.Endocr Relat Cancer. 2019 Nov;26(11):R611-R626. doi: 10.1530/ERC-19-0348. Endocr Relat Cancer. 2019. PMID: 31505466 Free PMC article. Review.
References
-
- Benabdallah NS, Bickmore WA. Regulatory Domains and Their Mechanisms. Cold Spring Harb Symp Quant Biol. 2015. Epub 2015/11/22. doi: 10.1101/sqb.2015.80.027268 . - DOI - PubMed
-
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152(6):1270–84. Epub 2013/03/19. doi: 10.1016/j.cell.2013.02.001 . - DOI - PubMed
-
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49(5):825–37. Epub 2013/03/12. doi: 10.1016/j.molcel.2013.01.038 ; PubMed Central PMCID: PMC3857148. - DOI - PMC - PubMed
-
- Dekker J, Misteli T. Long-Range Chromatin Interactions. Cold Spring Harb Perspect Biol. 2015;7(10). doi: 10.1101/cshperspect.a019356 . - DOI - PMC - PubMed
-
- Furlan-Magaril M, Varnai C, Nagano T, Fraser P. 3D genome architecture from populations to single cells. Curr Opin Genet Dev. 2015;31:36–41. doi: 10.1016/j.gde.2015.04.004 . - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

