Analysis of DNA methylation in chondrocytes in rats with knee osteoarthritis
- PMID: 28859619
- PMCID: PMC5579940
- DOI: 10.1186/s12891-017-1739-2
Analysis of DNA methylation in chondrocytes in rats with knee osteoarthritis
Abstract
Background: Knee osteoarthritis (KOA) is a degenerative knee disease commonly found in the ageing population. DNA methylation works with histone acetylation to participate in aging. Alterations of DNA methylation may involve the joint chondrocyte degeneration in KOA. The aim of this study is to detect DNA methylation changes in chondrocytes of rats with KOA.
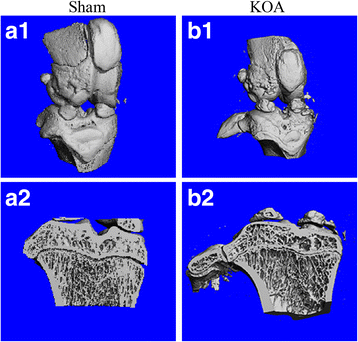
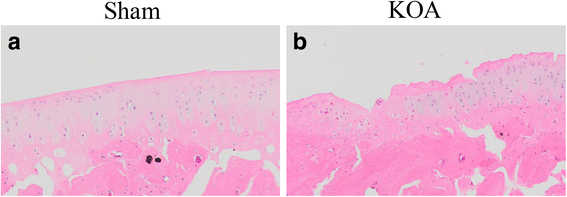
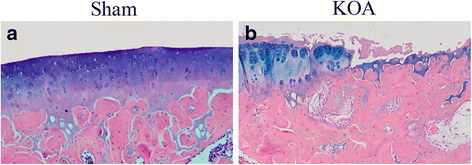
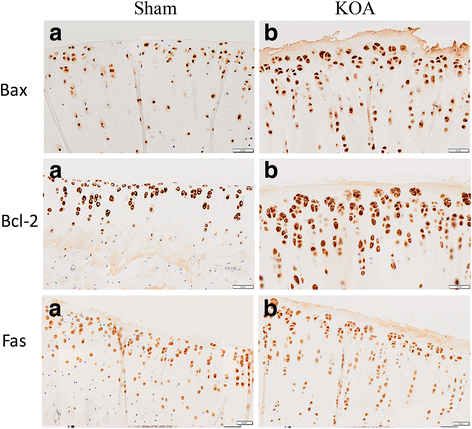
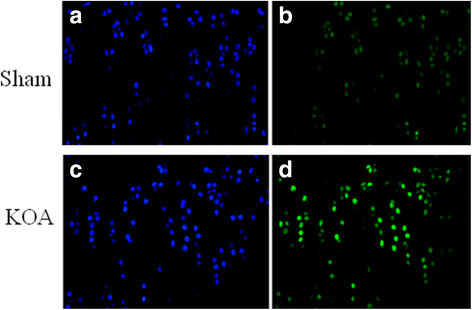
Methods: The rat KOA model was established with the Hulth method (n = 10), while rats receiving sham operation served as the control (n = 10). At 16 weeks after modeling, the knee joint tissue was collected from half of the rats in each group for Micro-CT scanning, Haematoxylin& Eosin (HE) staining, ABH/OG staining, immunohistochemistry for Bax, Bcl-2 and Fas, and TUNNEL staining. Meanwhile, the articular cartilage was collected from the other half to detect promoter methylation in target genes with the MethylTarget approach.
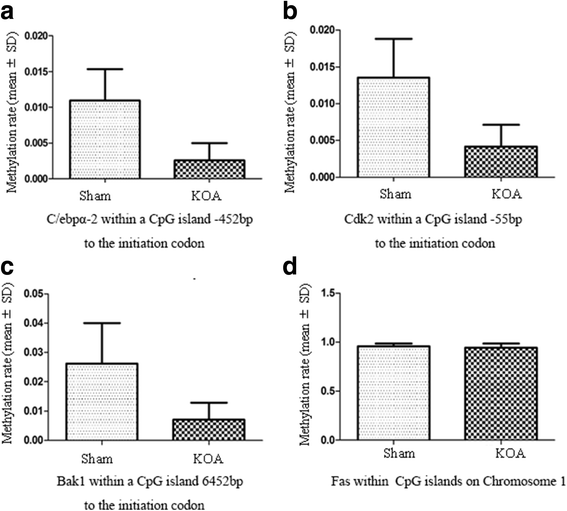
Results: Micro-CT scanning, HE staining, ABH/OG staining, immunohistochemistry, and TUNNEL staining all showed more severe cartilage injury in the KOA group than in the control group, indicating successful establishment of KOA model. The methylation rate in the KOA group was significantly decreased for C/ebpα-2 (within a CpG island -452 bp to the initiation codon on chromosome 1 91,363,511), Cdk2 (within a CpG island -55 bp to the initiation codon on chromosome 7 3,132,362), Bak1 (within a CpG island 6452 bp to the initiation codon on chromosome 20 5,622,277), and Fas (within a CpG island on the entire chromosome 1 gene), compared with the sham group (P = 0.005, 0.008, 0.022 and 0.027, respectively).
Conclusion: The chondrocyte apoptosis and significantly reduced methylation levels of C/ebpα-2, Cdk2, Bak1, and Fas may participate in the pathogenesis of KOA. However, the exact mechanisms remain to be determined.
Keywords: Bak; C/ebpα; Cdk2; Fas; Knee osteoarthritis; Methylation.
Conflict of interest statement
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Shanghai University of Traditional Chinese Medicine, China (No: SZY201507001). All procedures were in compliance with ethical guidelines for animal experiments. All efforts were made to minimize animal suffering.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interest with other people or any organizations.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Effects of Qi-Fang-Xi-Bi-Granules on Cartilage Morphology and C/ebpα Promoter Methylation in Rats with Knee Osteoarthritis.Evid Based Complement Alternat Med. 2018 Jan 28;2018:2074976. doi: 10.1155/2018/2074976. eCollection 2018. Evid Based Complement Alternat Med. 2018. PMID: 29670657 Free PMC article.
-
[Effects of inner-heating acupuncture on apoptosis of chondrocytes and expression of Caspase-3 and Caspase-9 in rats with knee osteoarthritis].Zhongguo Zhen Jiu. 2019 Apr 12;39(4):409-16. doi: 10.13703/j.0255-2930.2019.04.017. Zhongguo Zhen Jiu. 2019. PMID: 30957453 Chinese.
-
MicroRNA-199-3p up-regulation enhances chondrocyte proliferation and inhibits apoptosis in knee osteoarthritis via DNMT3A repression.Inflamm Res. 2021 Feb;70(2):171-182. doi: 10.1007/s00011-020-01430-1. Epub 2021 Jan 12. Inflamm Res. 2021. PMID: 33433641
-
Vitamin D and autophagy in knee osteoarthritis: A review.Int Immunopharmacol. 2023 Oct;123:110712. doi: 10.1016/j.intimp.2023.110712. Epub 2023 Jul 29. Int Immunopharmacol. 2023. PMID: 37523972 Review.
-
The role of oxidative stress in the development of knee osteoarthritis: A comprehensive research review.Front Mol Biosci. 2022 Sep 20;9:1001212. doi: 10.3389/fmolb.2022.1001212. eCollection 2022. Front Mol Biosci. 2022. PMID: 36203877 Free PMC article. Review.
Cited by
-
Osteoarthritis year in review 2018: genetics and epigenetics.Osteoarthritis Cartilage. 2019 Mar;27(3):371-377. doi: 10.1016/j.joca.2018.10.004. Epub 2018 Oct 25. Osteoarthritis Cartilage. 2019. PMID: 30808485 Free PMC article. Review.
-
Folate treatment of pregnant rat dams abolishes metabolic effects in female offspring induced by a paternal pre-conception unhealthy diet.Diabetologia. 2018 Aug;61(8):1862-1876. doi: 10.1007/s00125-018-4635-x. Epub 2018 May 18. Diabetologia. 2018. PMID: 29777263
-
Integrated Analysis of DNA Methylation and Gene Expression Profiles in a Rat Model of Osteoarthritis.Int J Mol Sci. 2024 Jan 2;25(1):594. doi: 10.3390/ijms25010594. Int J Mol Sci. 2024. PMID: 38203768 Free PMC article.
-
The Effect of Intra-articular versus Intravenous Injection of Mesenchymal Stem Cells on Experimentally-Induced Knee Joint Osteoarthritis.J Microsc Ultrastruct. 2020 May 20;9(1):31-38. doi: 10.4103/JMAU.JMAU_2_20. eCollection 2021 Jan-Mar. J Microsc Ultrastruct. 2020. PMID: 33850710 Free PMC article.
-
Polyglutamine-expanded ataxin3 alter specific gene expressions through changing DNA methylation status in SCA3/MJD.Aging (Albany NY). 2020 Dec 19;13(3):3680-3698. doi: 10.18632/aging.202331. Epub 2020 Dec 19. Aging (Albany NY). 2020. PMID: 33411688 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

