Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4
- PMID: 28859267
- PMCID: PMC5583414
- DOI: 10.4142/jvs.2017.18.S1.269
Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4
Abstract
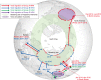
Novel subtypes of Asian-origin (Goose/Guangdong lineage) H5 highly pathogenic avian influenza (HPAI) viruses belonging to clade 2.3.4, such as H5N2, H5N5, H5N6, and H5N8, have been identified in China since 2008 and have since evolved into four genetically distinct clade 2.3.4.4 groups (A-D). Since 2014, HPAI clade 2.3.4.4 viruses have spread rapidly via migratory wild aquatic birds and have evolved through reassortment with prevailing local low pathogenicity avian influenza viruses. Group A H5N8 viruses and its reassortant viruses caused outbreaks in wide geographic regions (Asia, Europe, and North America) during 2014-2015. Novel reassortant Group B H5N8 viruses caused outbreaks in Asia, Europe, and Africa during 2016-2017. Novel reassortant Group C H5N6 viruses caused outbreaks in Korea and Japan during the 2016-2017 winter season. Group D H5N6 viruses caused outbreaks in China and Vietnam. A wide range of avian species, including wild and domestic waterfowl, domestic poultry, and even zoo birds, seem to be permissive for infection by and/or transmission of clade 2.3.4.4 HPAI viruses. Further, compared to previous H5N1 HPAI viruses, these reassortant viruses show altered pathogenicity in birds. In this review, we discuss the evolution, global spread, and pathogenicity of H5 clade 2.3.4.4 HPAI viruses.
Keywords: epidemiology; influenza in birds; poultry; transmission; virulence.
Conflict of interest statement
Figures
Similar articles
-
Genetic diversity of highly pathogenic avian influenza H5N6 and H5N8 viruses in poultry markets in Guangdong, China, 2020-2022.J Virol. 2025 Jan 31;99(1):e0114524. doi: 10.1128/jvi.01145-24. Epub 2024 Dec 4. J Virol. 2025. PMID: 39629997 Free PMC article.
-
Pathogenesis and Transmission of Novel Highly Pathogenic Avian Influenza H5N2 and H5N8 Viruses in Ferrets and Mice.J Virol. 2015 Oct;89(20):10286-93. doi: 10.1128/JVI.01438-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223637 Free PMC article.
-
The genetics of highly pathogenic avian influenza viruses of subtype H5 in Germany, 2006-2020.Transbound Emerg Dis. 2021 May;68(3):1136-1150. doi: 10.1111/tbed.13843. Epub 2020 Sep 29. Transbound Emerg Dis. 2021. PMID: 32964686 Review.
-
Pathobiology of Clade 2.3.4.4 H5Nx High-Pathogenicity Avian Influenza Virus Infections in Minor Gallinaceous Poultry Supports Early Backyard Flock Introductions in the Western United States in 2014-2015.J Virol. 2017 Oct 13;91(21):e00960-17. doi: 10.1128/JVI.00960-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28794040 Free PMC article.
-
Emergence and dissemination of clade 2.3.4.4 H5Nx influenza viruses-how is the Asian HPAI H5 lineage maintained.Curr Opin Virol. 2016 Feb;16:158-163. doi: 10.1016/j.coviro.2016.02.005. Epub 2016 Mar 15. Curr Opin Virol. 2016. PMID: 26991931 Review.
Cited by
-
Genetic characterization of an H5N6 avian influenza virus with multiple origins from a chicken in southern China, October 2019.BMC Vet Res. 2021 May 28;17(1):200. doi: 10.1186/s12917-021-02903-z. BMC Vet Res. 2021. PMID: 34049549 Free PMC article.
-
Avian influenza overview November 2019- February2020.EFSA J. 2020 Mar 31;18(3):e06096. doi: 10.2903/j.efsa.2020.6096. eCollection 2020 Mar. EFSA J. 2020. PMID: 32874270 Free PMC article.
-
The roles of migratory and resident birds in local avian influenza infection dynamics.J Appl Ecol. 2018 Nov;55(6):2963-2975. doi: 10.1111/1365-2664.13154. Epub 2018 Mar 26. J Appl Ecol. 2018. PMID: 30337766 Free PMC article.
-
Monoclonal antibody targeting a novel linear epitope on nucleoprotein confers pan-reactivity to influenza A virus.Appl Microbiol Biotechnol. 2023 Apr;107(7-8):2437-2450. doi: 10.1007/s00253-023-12433-3. Epub 2023 Feb 23. Appl Microbiol Biotechnol. 2023. PMID: 36820898 Free PMC article.
-
Efficacy of a Recombinant Turkey Herpesvirus AI (H5) Vaccine in Preventing Transmission of Heterologous Highly Pathogenic H5N8 Clade 2.3.4.4b Challenge Virus in Commercial Broilers and Layer Pullets.J Immunol Res. 2018 Nov 21;2018:3143189. doi: 10.1155/2018/3143189. eCollection 2018. J Immunol Res. 2018. PMID: 30584541 Free PMC article.
References
-
- Berhane Y, Kobasa D, Embury-Hyatt C, Pickering B, Babiuk S, Joseph T, Bowes V, Suderman M, Leung A, Cottam-Birt C, Hisanaga T, Pasick J. Pathobiological characterization of a novel reassortant highly pathogenic H5N1 virus isolated in British Columbia, Canada, 2015. Sci Rep. 2016;6:23380. - PMC - PubMed
-
- Bertran K, Swayne DE, Pantin-Jackwood MJ, Kapczynski DR, Spackman E, Suarez DL. Lack of chicken adaptation of newly emergent Eurasian H5N8 and reassortant H5N2 high pathogenicity avian influenza viruses in the U.S. is consistent with restricted poultry outbreaks in the Pacific flyway during 2014-2015. Virology. 2016;494:190–197. - PubMed
-
- Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, Quan C, Liu J, Wu J, Yin R, Zhao L, Li M, Ding Z, Zou R, Xu W, Li H, Wang H, Tian K, Fu G, Huang Y, Shestopalov A, Li S, Xu B, Yu H, Luo T, Lu L, Xu X, Luo Y, Liu Y, Shi W, Liu D, Gao GF. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe. 2016;20:810–821. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical