Extracellular superoxide dismutase and its role in cancer
- PMID: 28842347
- PMCID: PMC5685559
- DOI: 10.1016/j.freeradbiomed.2017.08.013
Extracellular superoxide dismutase and its role in cancer
Abstract
Reactive oxygen species (ROS) are increasingly recognized as critical determinants of cellular signaling and a strict balance of ROS levels must be maintained to ensure proper cellular function and survival. Notably, ROS is increased in cancer cells. The superoxide dismutase family plays an essential physiological role in mitigating deleterious effects of ROS. Due to the compartmentalization of ROS signaling, EcSOD, the only superoxide dismutase in the extracellular space, has unique characteristics and functions in cellular signal transduction. In comparison to the other two intracellular SODs, EcSOD is a relatively new comer in terms of its tumor suppressive role in cancer and the mechanisms involved are less well understood. Nevertheless, the degree of differential expression of this extracellular antioxidant in cancer versus normal cells/tissues is more pronounced and prevalent than the other SODs. A significant association of low EcSOD expression with reduced cancer patient survival further suggests that loss of extracellular redox regulation promotes a conducive microenvironment that favors cancer progression. The vast array of mechanisms reported in mediating deregulation of EcSOD expression, function, and cellular distribution also supports that loss of this extracellular antioxidant provides a selective advantage to cancer cells. Moreover, overexpression of EcSOD inhibits tumor growth and metastasis, indicating a role as a tumor suppressor. This review focuses on the current understanding of the mechanisms of deregulation and tumor suppressive function of EcSOD in cancer.
Keywords: Cancer; EcSOD; Epigenetic; Heparin binding domain; Loss of heterozygosity; Metastasis; Oxidative tumor microenvironment; Reactive oxygen species; Recurrence; Relapse free survival; SOD3; Single nucleotide polymorphism; Tumor suppressor; microRNA-21.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no potential conflicts of interest
Figures

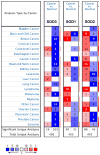
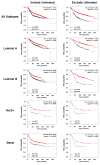
Similar articles
-
Expanding roles of superoxide dismutases in cell regulation and cancer.Drug Discov Today. 2016 Jan;21(1):143-149. doi: 10.1016/j.drudis.2015.10.001. Epub 2015 Oct 19. Drug Discov Today. 2016. PMID: 26475962 Free PMC article. Review.
-
[Regulation of Extracellular Redox Homeostasis in Tumor Microenvironment].Yakugaku Zasshi. 2019;139(9):1139-1144. doi: 10.1248/yakushi.19-00128. Yakugaku Zasshi. 2019. PMID: 31474628 Review. Japanese.
-
Essential role of extracellular SOD in reparative neovascularization induced by hindlimb ischemia.Circ Res. 2007 Aug 17;101(4):409-19. doi: 10.1161/CIRCRESAHA.107.153791. Epub 2007 Jun 29. Circ Res. 2007. PMID: 17601801
-
Extracellular superoxide dismutase suppresses hypoxia-inducible factor-1α in pancreatic cancer.Free Radic Biol Med. 2014 Apr;69:357-66. doi: 10.1016/j.freeradbiomed.2014.02.002. Epub 2014 Feb 7. Free Radic Biol Med. 2014. PMID: 24509158 Free PMC article.
-
Imperative connotation of SODs in cancer: Emerging targets and multifactorial role of action.IUBMB Life. 2024 Sep;76(9):592-613. doi: 10.1002/iub.2821. Epub 2024 Apr 10. IUBMB Life. 2024. PMID: 38600696 Review.
Cited by
-
Evaluation of antioxidant network proteins as novel prognostic biomarkers for head and neck cancer patients.Oral Oncol. 2020 Dec;111:104949. doi: 10.1016/j.oraloncology.2020.104949. Epub 2020 Aug 12. Oral Oncol. 2020. PMID: 32801084 Free PMC article.
-
The Involvement of the Oxidative Stress Status in Cancer Pathology: A Double View on the Role of the Antioxidants.Oxid Med Cell Longev. 2021 Aug 5;2021:9965916. doi: 10.1155/2021/9965916. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 34394838 Free PMC article. Review.
-
Dietary Supplementation of Chestnut Tannins in Prepartum Dairy Cows Improves Antioxidant Defense Mechanisms Interacting with Thyroid Status.Metabolites. 2023 Feb 24;13(3):334. doi: 10.3390/metabo13030334. Metabolites. 2023. PMID: 36984774 Free PMC article.
-
Protective Effect of Hesperidin on the Oxidative Stress Induced by an Exhausting Exercise in Intensively Trained Rats.Nutrients. 2019 Apr 4;11(4):783. doi: 10.3390/nu11040783. Nutrients. 2019. PMID: 30987366 Free PMC article.
-
Nano Uncaria gambir as Chemopreventive Agent Against Breast Cancer.Int J Nanomedicine. 2023 Aug 3;18:4471-4484. doi: 10.2147/IJN.S403385. eCollection 2023. Int J Nanomedicine. 2023. PMID: 37555190 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

