Expression and Subcellular Localization of the Kaposi's Sarcoma-Associated Herpesvirus K15P Protein during Latency and Lytic Reactivation in Primary Effusion Lymphoma Cells
- PMID: 28835496
- PMCID: PMC5640876
- DOI: 10.1128/JVI.01370-17
Expression and Subcellular Localization of the Kaposi's Sarcoma-Associated Herpesvirus K15P Protein during Latency and Lytic Reactivation in Primary Effusion Lymphoma Cells
Abstract
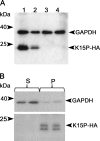
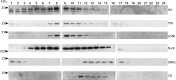
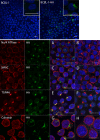
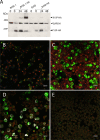
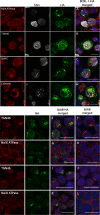
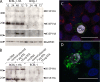
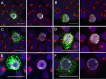
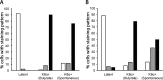
The K15P membrane protein of Kaposi's sarcoma-associated herpesvirus (KSHV) interacts with multiple cellular signaling pathways and is thought to play key roles in KSHV-associated endothelial cell angiogenesis, regulation of B-cell receptor (BCR) signaling, and the survival, activation, and proliferation of BCR-negative primary effusion lymphoma (PEL) cells. Although full-length K15P is ∼45 kDa, numerous lower-molecular-weight forms of the protein exist as a result of differential splicing and poorly characterized posttranslational processing. K15P has been reported to localize to numerous subcellular organelles in heterologous expression studies, but there are limited data concerning the sorting of K15P in KSHV-infected cells. The relationships between the various molecular weight forms of K15P, their subcellular distribution, and how these may differ in latent and lytic KSHV infections are poorly understood. Here we report that a cDNA encoding a full-length, ∼45-kDa K15P reporter protein is expressed as an ∼23- to 24-kDa species that colocalizes with the trans-Golgi network (TGN) marker TGN46 in KSHV-infected PEL cells. Following lytic reactivation by sodium butyrate, the levels of the ∼23- to 24-kDa protein diminish, and the full-length, ∼45-kDa K15P protein accumulates. This is accompanied by apparent fragmentation of the TGN and redistribution of K15P to a dispersed peripheral location. Similar results were seen when lytic reactivation was stimulated by the KSHV protein replication and transcription activator (RTA) and during spontaneous reactivation. We speculate that expression of different molecular weight forms of K15P in distinct cellular locations reflects the alternative demands placed upon the protein in the latent and lytic phases.IMPORTANCE The K15P protein of Kaposi's sarcoma-associated herpesvirus (KSHV) is thought to play key roles in disease, including KSHV-associated angiogenesis and the survival and growth of primary effusion lymphoma (PEL) cells. The protein exists in multiple molecular weight forms, and its intracellular trafficking is poorly understood. Here we demonstrate that the molecular weight form of a reporter K15P molecule and its intracellular distribution change when KSHV switches from its latent (quiescent) phase to the lytic, infectious state. We speculate that expression of different molecular weight forms of K15P in distinct cellular locations reflects the alternative demands placed upon the protein in the viral latent and lytic stages.
Keywords: K15P; Kaposi's sarcoma-associated herpesvirus; latent and lytic infection; subcellular localization.
Copyright © 2017 American Society for Microbiology.
Figures
Similar articles
-
ARID3B: a Novel Regulator of the Kaposi's Sarcoma-Associated Herpesvirus Lytic Cycle.J Virol. 2016 Sep 29;90(20):9543-55. doi: 10.1128/JVI.03262-15. Print 2016 Oct 15. J Virol. 2016. PMID: 27512077 Free PMC article.
-
Activated Nrf2 Interacts with Kaposi's Sarcoma-Associated Herpesvirus Latency Protein LANA-1 and Host Protein KAP1 To Mediate Global Lytic Gene Repression.J Virol. 2015 Aug;89(15):7874-92. doi: 10.1128/JVI.00895-15. Epub 2015 May 20. J Virol. 2015. PMID: 25995248 Free PMC article.
-
Degradation of TRIM32 is induced by RTA for Kaposi's sarcoma-associated herpesvirus lytic replication.J Virol. 2024 Jun 13;98(6):e0000524. doi: 10.1128/jvi.00005-24. Epub 2024 May 8. J Virol. 2024. PMID: 38717113 Free PMC article.
-
Regulation of KSHV Latency and Lytic Reactivation.Viruses. 2020 Sep 17;12(9):1034. doi: 10.3390/v12091034. Viruses. 2020. PMID: 32957532 Free PMC article. Review.
-
Cyclooxygenase-2-prostaglandin E2-eicosanoid receptor inflammatory axis: a key player in Kaposi's sarcoma-associated herpes virus associated malignancies.Transl Res. 2013 Aug;162(2):77-92. doi: 10.1016/j.trsl.2013.03.004. Epub 2013 Apr 6. Transl Res. 2013. PMID: 23567332 Free PMC article. Review.
Cited by
-
Kaposi's Sarcoma-Associated Herpesvirus Nonstructural Membrane Protein pK15 Recruits the Class II Phosphatidylinositol 3-Kinase PI3K-C2α To Activate Productive Viral Replication.J Virol. 2018 Aug 16;92(17):e00544-18. doi: 10.1128/JVI.00544-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29950425 Free PMC article.
-
Viral Causality of Human Cancer and Potential Roles of Human Endogenous Retroviruses in the Multi-Omics Era: An Evolutionary Epidemiology Review.Front Oncol. 2021 Oct 29;11:687631. doi: 10.3389/fonc.2021.687631. eCollection 2021. Front Oncol. 2021. PMID: 34778024 Free PMC article. Review.
References
-
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276–1280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

