Genetic and codon usage bias analyses of polymerase genes of equine influenza virus and its relation to evolution
- PMID: 28830350
- PMCID: PMC5568313
- DOI: 10.1186/s12864-017-4063-1
Genetic and codon usage bias analyses of polymerase genes of equine influenza virus and its relation to evolution
Abstract
Background: Equine influenza is a major health problem of equines worldwide. The polymerase genes of influenza virus have key roles in virus replication, transcription, transmission between hosts and pathogenesis. Hence, the comprehensive genetic and codon usage bias of polymerase genes of equine influenza virus (EIV) were analyzed to elucidate the genetic and evolutionary relationships in a novel perspective.
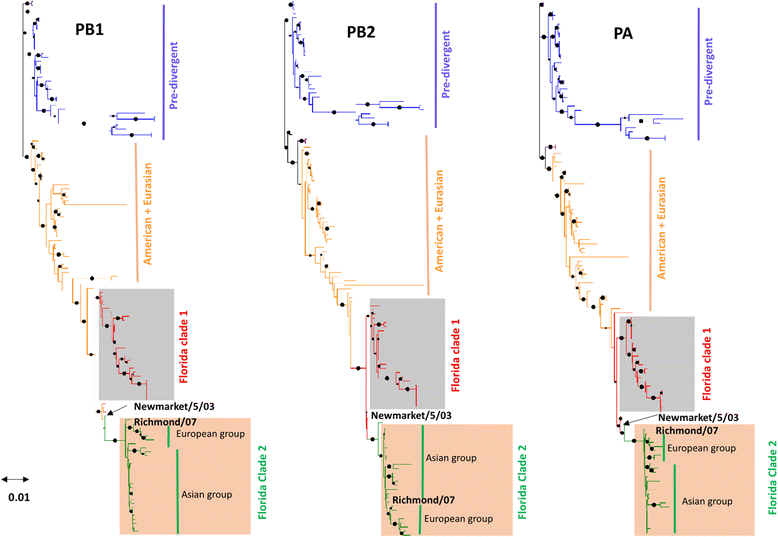
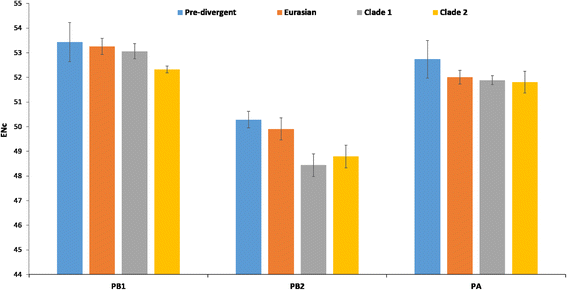
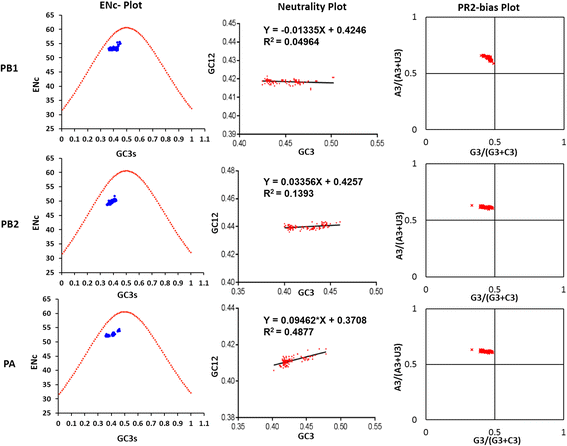
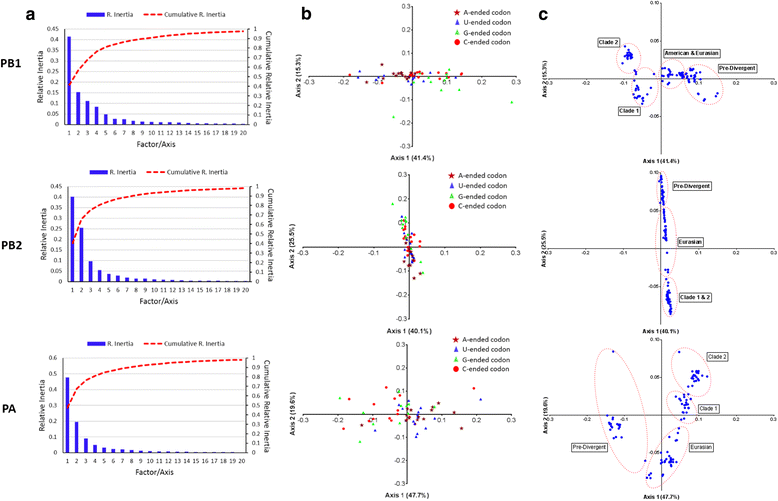
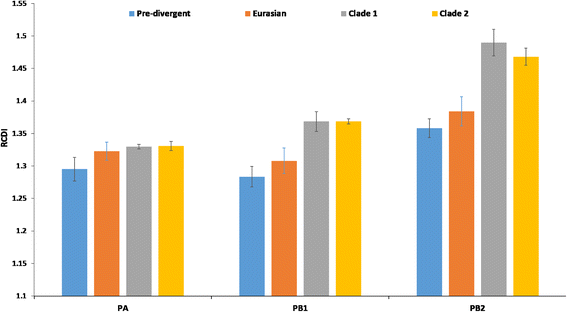
Results: The group - specific consensus amino acid substitutions were identified in all polymerase genes of EIVs that led to divergence of EIVs into various clades. The consistent amino acid changes were also detected in the Florida clade 2 EIVs circulating in Europe and Asia since 2007. To study the codon usage patterns, a total of 281,324 codons of polymerase genes of EIV H3N8 isolates from 1963 to 2015 were systemically analyzed. The polymerase genes of EIVs exhibit a weak codon usage bias. The ENc-GC3s and Neutrality plots indicated that natural selection is the major influencing factor of codon usage bias, and that the impact of mutation pressure is comparatively minor. The methods for estimating host imposed translation pressure suggested that the polymerase acidic (PA) gene seems to be under less translational pressure compared to polymerase basic 1 (PB1) and polymerase basic 2 (PB2) genes. The multivariate statistical analysis of polymerase genes divided EIVs into four evolutionary diverged clusters - Pre-divergent, Eurasian, Florida sub-lineage 1 and 2.
Conclusions: Various lineage specific amino acid substitutions observed in all polymerase genes of EIVs and especially, clade 2 EIVs underwent major variations which led to the emergence of a phylogenetically distinct group of EIVs originating from Richmond/1/07. The codon usage bias was low in all the polymerase genes of EIVs that was influenced by the multiple factors such as the nucleotide compositions, mutation pressure, aromaticity and hydropathicity. However, natural selection was the major influencing factor in defining the codon usage patterns and evolution of polymerase genes of EIVs.
Keywords: Codon usage bias; Equine influenza virus; Evolution; H3N8; Polymerase genes.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Revelation of Influencing Factors in Overall Codon Usage Bias of Equine Influenza Viruses.PLoS One. 2016 Apr 27;11(4):e0154376. doi: 10.1371/journal.pone.0154376. eCollection 2016. PLoS One. 2016. PMID: 27119730 Free PMC article.
-
Equine Influenza Virus in Asia: Phylogeographic Pattern and Molecular Features Reveal Circulation of an Autochthonous Lineage.J Virol. 2019 Jun 14;93(13):e00116-19. doi: 10.1128/JVI.00116-19. Print 2019 Jul 1. J Virol. 2019. PMID: 31019053 Free PMC article.
-
Genetic analysis of the PB1-F2 gene of equine influenza virus.Virus Genes. 2013 Oct;47(2):250-8. doi: 10.1007/s11262-013-0935-x. Epub 2013 Jun 19. Virus Genes. 2013. PMID: 23780220
-
Equine Influenza Virus: An Old Known Enemy in the Americas.Vaccines (Basel). 2022 Oct 14;10(10):1718. doi: 10.3390/vaccines10101718. Vaccines (Basel). 2022. PMID: 36298583 Free PMC article. Review.
-
Gene expression and molecular evolution.Curr Opin Genet Dev. 2001 Dec;11(6):660-6. doi: 10.1016/s0959-437x(00)00250-1. Curr Opin Genet Dev. 2001. PMID: 11682310 Review.
Cited by
-
Origin and Evolution of H1N1/pdm2009: A Codon Usage Perspective.Front Microbiol. 2020 Jul 14;11:1615. doi: 10.3389/fmicb.2020.01615. eCollection 2020. Front Microbiol. 2020. PMID: 32760376 Free PMC article.
-
Reconstruction of the Evolutionary Origin, Phylodynamics, and Phylogeography of the Porcine Circovirus Type 3.Front Microbiol. 2022 May 18;13:898212. doi: 10.3389/fmicb.2022.898212. eCollection 2022. Front Microbiol. 2022. PMID: 35663871 Free PMC article.
-
Genetic and evolutionary analysis of emerging H3N2 canine influenza virus.Emerg Microbes Infect. 2018 Apr 25;7(1):73. doi: 10.1038/s41426-018-0079-0. Emerg Microbes Infect. 2018. PMID: 29691381 Free PMC article.
-
Analysis of the Codon Usage Pattern of HA and NA Genes of H7N9 Influenza A Virus.Int J Mol Sci. 2020 Sep 27;21(19):7129. doi: 10.3390/ijms21197129. Int J Mol Sci. 2020. PMID: 32992529 Free PMC article.
-
Genetic Evolution and Molecular Selection of the HE Gene of Influenza C Virus.Viruses. 2019 Feb 19;11(2):167. doi: 10.3390/v11020167. Viruses. 2019. PMID: 30791465 Free PMC article.
References
-
- Bryant NA, Rash AS, Russell CA, Ross J, Cooke A, Bowman S, MacRae S, Lewis NS, Paillot R, Zanoni R, Meier H, Griffiths LA, Daly JM, Tiwari A, Chambers TM, Newton JR, Elton DM. Antigenic and genetic variations in European and north American equine influenza virus strains (H3N8) isolated from 2006 to 2007. Vet Microbiol. 2009;138:41–52. doi: 10.1016/j.vetmic.2009.03.004. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

