Analysis of AgoshRNA maturation and loading into Ago2
- PMID: 28809941
- PMCID: PMC5557517
- DOI: 10.1371/journal.pone.0183269
Analysis of AgoshRNA maturation and loading into Ago2
Abstract
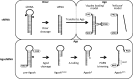
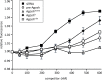
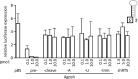
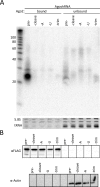
The RNA interference (RNAi) pathway was recently expanded by the discovery of multiple alternative pathways for processing of natural microRNA (miRNA) and man-made short hairpin RNA (shRNA) molecules. One non-canonical pathway bypasses Dicer cleavage and requires instead processing by Argonaute2 (Ago2), which also executes the subsequent silencing step. We named these molecules AgoshRNA, which generate only a single active RNA strand and thus avoid off-target effects that can be induced by the passenger strand of a regular shRNA. Previously, we characterized AgoshRNA processing by deep sequencing and demonstrated that-after Ago2 cleavage-AgoshRNAs acquire a short 3' tail of 1-3 A-nucleotides and are subsequently trimmed, likely by the poly(A)-specific ribonuclease (PARN). As a result, the mature single-stranded AgoshRNA may dock more stably into Ago2. Here we set out to analyze the activity of different synthetic AgoshRNA processing intermediates. Ago2 was found to bind preferentially to partially single-stranded AgoshRNA in vitro. In contrast, only the double-stranded AgoshRNA precursor associated with Ago2 in cells, correlating with efficient intracellular processing and reporter knockdown activity. These results suggest the presence of a cellular co-factor involved in AgoshRNA loading into Ago2 in vivo. We also demonstrate specific AgoshRNA loading in Ago2, but not Ago1/3/4, thus further reducing unwanted side effects.
Conflict of interest statement
Figures


Similar articles
-
Dicer-independent processing of small RNA duplexes: mechanistic insights and applications.Nucleic Acids Res. 2017 Oct 13;45(18):10369-10379. doi: 10.1093/nar/gkx779. Nucleic Acids Res. 2017. PMID: 28977573 Free PMC article. Review.
-
Silencing of HIV-1 by AgoshRNA molecules.Gene Ther. 2017 Aug;24(8):453-461. doi: 10.1038/gt.2017.44. Epub 2017 May 29. Gene Ther. 2017. PMID: 28553929
-
Mechanistic insights on the Dicer-independent AGO2-mediated processing of AgoshRNAs.RNA Biol. 2015;12(1):92-100. doi: 10.1080/15476286.2015.1017204. RNA Biol. 2015. PMID: 25826416 Free PMC article.
-
Toward optimization of AgoshRNA molecules that use a non-canonical RNAi pathway: variations in the top and bottom base pairs.RNA Biol. 2015;12(4):447-56. doi: 10.1080/15476286.2015.1022024. RNA Biol. 2015. PMID: 25747107 Free PMC article.
-
AGO2 and its partners: a silencing complex, a chromatin modulator, and new features.Crit Rev Biochem Mol Biol. 2020 Feb;55(1):33-53. doi: 10.1080/10409238.2020.1738331. Epub 2020 Mar 13. Crit Rev Biochem Mol Biol. 2020. PMID: 32164444 Review.
Cited by
-
Subretinal AAV delivery of RNAi-therapeutics targeting VEGFA reduces choroidal neovascularization in a large animal model.Mol Ther Methods Clin Dev. 2024 Mar 22;32(2):101242. doi: 10.1016/j.omtm.2024.101242. eCollection 2024 Jun 13. Mol Ther Methods Clin Dev. 2024. PMID: 38605811 Free PMC article.
-
Cytoplasmic cleavage of IMPA1 3' UTR is necessary for maintaining axon integrity.Cell Rep. 2021 Feb 23;34(8):108778. doi: 10.1016/j.celrep.2021.108778. Cell Rep. 2021. PMID: 33626357 Free PMC article.
-
miRNA-encapsulated abiotic materials and biovectors for cutaneous and oral wound healing: Biogenesis, mechanisms, and delivery nanocarriers.Bioeng Transl Med. 2022 Jun 28;8(1):e10343. doi: 10.1002/btm2.10343. eCollection 2023 Jan. Bioeng Transl Med. 2022. PMID: 36684081 Free PMC article. Review.
-
Dicer-independent processing of small RNA duplexes: mechanistic insights and applications.Nucleic Acids Res. 2017 Oct 13;45(18):10369-10379. doi: 10.1093/nar/gkx779. Nucleic Acids Res. 2017. PMID: 28977573 Free PMC article. Review.
-
Accumulation of AGO2 Facilitates Tumorigenesis of Human Hepatocellular Carcinoma.Biomed Res Int. 2020 Apr 30;2020:1631843. doi: 10.1155/2020/1631843. eCollection 2020. Biomed Res Int. 2020. PMID: 32420319 Free PMC article.
References
-
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002 ; PubMed Central PMCID: PMC3794896. - DOI - PMC - PubMed
-
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–5. doi: 10.1038/nature03049 . - DOI - PubMed
-
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–40. doi: 10.1038/nature03120 . - DOI - PubMed
-
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21(21):5864–74. doi: 10.1093/emboj/cdf578 ; PubMed Central PMCID: PMC131075. - DOI - PMC - PubMed
-
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21(21):5875–85. doi: 10.1093/emboj/cdf582 ; PubMed Central PMCID: PMC131079. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

