Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1
- PMID: 28781234
- PMCID: PMC5957536
- DOI: 10.1016/j.molcel.2017.06.035
Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1
Abstract
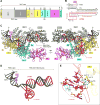
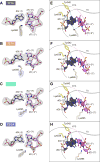
The RNA-guided Cpf1 (also known as Cas12a) nuclease associates with a CRISPR RNA (crRNA) and cleaves the double-stranded DNA target complementary to the crRNA guide. The two Cpf1 orthologs from Acidaminococcus sp. (AsCpf1) and Lachnospiraceae bacterium (LbCpf1) have been harnessed for eukaryotic genome editing. Cpf1 requires a specific nucleotide sequence, called a protospacer adjacent motif (PAM), for target recognition. Besides the canonical TTTV PAM, Cpf1 recognizes suboptimal C-containing PAMs. Here, we report four crystal structures of LbCpf1 in complex with the crRNA and its target DNA containing either TTTA, TCTA, TCCA, or CCCA as the PAM. These structures revealed that, depending on the PAM sequences, LbCpf1 undergoes conformational changes to form altered interactions with the PAM-containing DNA duplexes, thereby achieving the relaxed PAM recognition. Collectively, the present structures advance our mechanistic understanding of the PAM-dependent, crRNA-guided DNA cleavage by the Cpf1 family nucleases.
Keywords: CRISPR-Cas; Cas12a; Cpf1; crystal structure; protospacer adjacent motif.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1.Mol Cell. 2017 Jul 6;67(1):139-147.e2. doi: 10.1016/j.molcel.2017.04.019. Epub 2017 Jun 6. Mol Cell. 2017. PMID: 28595896 Free PMC article.
-
Structural Variation of Type I-F CRISPR RNA Guided DNA Surveillance.Mol Cell. 2017 Aug 17;67(4):622-632.e4. doi: 10.1016/j.molcel.2017.06.036. Epub 2017 Aug 3. Mol Cell. 2017. PMID: 28781236
-
Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a).Proc Natl Acad Sci U S A. 2018 May 22;115(21):5444-5449. doi: 10.1073/pnas.1718686115. Epub 2018 May 7. Proc Natl Acad Sci U S A. 2018. PMID: 29735714 Free PMC article.
-
Methods for decoding Cas9 protospacer adjacent motif (PAM) sequences: A brief overview.Methods. 2017 May 15;121-122:3-8. doi: 10.1016/j.ymeth.2017.03.006. Epub 2017 Mar 24. Methods. 2017. PMID: 28344037 Review.
-
The Conspicuity of CRISPR-Cpf1 System as a Significant Breakthrough in Genome Editing.Curr Microbiol. 2018 Jan;75(1):107-115. doi: 10.1007/s00284-017-1406-8. Epub 2017 Nov 30. Curr Microbiol. 2018. PMID: 29189942 Review.
Cited by
-
New PAM Improves the Single-Base Specificity of crRNA-Guided LbCas12a Nuclease.Life (Basel). 2022 Nov 18;12(11):1927. doi: 10.3390/life12111927. Life (Basel). 2022. PMID: 36431062 Free PMC article.
-
Endogenous CRISPR-Cas System-Based Genome Editing and Antimicrobials: Review and Prospects.Front Microbiol. 2019 Oct 25;10:2471. doi: 10.3389/fmicb.2019.02471. eCollection 2019. Front Microbiol. 2019. PMID: 31708910 Free PMC article. Review.
-
Characterization of Cas12a nucleases reveals diverse PAM profiles between closely-related orthologs.Nucleic Acids Res. 2020 Jun 4;48(10):5624-5638. doi: 10.1093/nar/gkaa272. Nucleic Acids Res. 2020. PMID: 32329776 Free PMC article.
-
Interfering with retrotransposition by two types of CRISPR effectors: Cas12a and Cas13a.Cell Discov. 2020 May 19;6:30. doi: 10.1038/s41421-020-0164-0. eCollection 2020. Cell Discov. 2020. PMID: 32435507 Free PMC article.
-
A Cas12a ortholog with stringent PAM recognition followed by low off-target editing rates for genome editing.Genome Biol. 2020 Mar 25;21(1):78. doi: 10.1186/s13059-020-01989-2. Genome Biol. 2020. PMID: 32213191 Free PMC article.
References
-
- Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. - PubMed
-
- Dong D, Ren K, Qiu X, Zheng J, Guo M, Guan X, Liu H, Li N, Zhang B, Yang D, et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016;532:522–526. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

