RIP-Seq Suggests Translational Regulation by L7Ae in Archaea
- PMID: 28765217
- PMCID: PMC5539422
- DOI: 10.1128/mBio.00730-17
RIP-Seq Suggests Translational Regulation by L7Ae in Archaea
Abstract
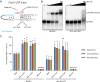
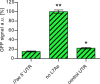
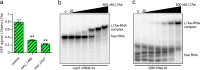
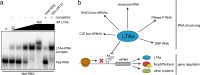
L7Ae is a universal archaeal protein that recognizes and stabilizes kink-turn (k-turn) motifs in RNA substrates. These structural motifs are widespread in nature and are found in many functional RNA species, including ribosomal RNAs. Synthetic biology approaches utilize L7Ae/k-turn interactions to control gene expression in eukaryotes. Here, we present results of comprehensive RNA immunoprecipitation sequencing (RIP-Seq) analysis of genomically tagged L7Ae from the hyperthermophilic archaeon Sulfolobus acidocaldarius A large set of interacting noncoding RNAs was identified. In addition, several mRNAs, including the l7ae transcript, were found to contain k-turn motifs that facilitate L7Ae binding. In vivo studies showed that L7Ae autoregulates the translation of its mRNA by binding to a k-turn motif present in the 5' untranslated region (UTR). A green fluorescent protein (GFP) reporter system was established in Escherichia coli and verified conservation of L7Ae-mediated feedback regulation in Archaea Mobility shift assays confirmed binding to a k-turn in the transcript of nop5-fibrillarin, suggesting that the expression of all C/D box sRNP core proteins is regulated by L7Ae. These studies revealed that L7Ae-mediated gene regulation evolved in archaeal organisms, generating new tools for the modulation of synthetic gene circuits in bacteria.IMPORTANCE L7Ae is an essential archaeal protein that is known to structure ribosomal RNAs and small RNAs (sRNAs) by binding to their kink-turn motifs. Here, we utilized RIP-Seq methodology to achieve a first global analysis of RNA substrates for L7Ae. Several novel interactions with noncoding RNA molecules (e.g., with the universal signal recognition particle RNA) were discovered. In addition, L7Ae was found to bind to mRNAs, including its own transcript's 5' untranslated region. This feedback-loop control is conserved in most archaea and was incorporated into a reporter system that was utilized to control gene expression in bacteria. These results demonstrate that L7Ae-mediated gene regulation evolved originally in archaeal organisms. The feedback-controlled reporter gene system can easily be adapted for synthetic biology approaches that require strict gene expression control.
Keywords: Archaea; RNA binding proteins; RNA structure; gene regulation.
Copyright © 2017 Daume et al.
Figures



Similar articles
-
Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea.Nucleic Acids Res. 2003 Feb 1;31(3):869-77. doi: 10.1093/nar/gkg175. Nucleic Acids Res. 2003. PMID: 12560482 Free PMC article.
-
The role of RNA structure in translational regulation by L7Ae protein in archaea.RNA. 2019 Jan;25(1):60-69. doi: 10.1261/rna.068510.118. Epub 2018 Oct 16. RNA. 2019. PMID: 30327333 Free PMC article.
-
Signature amino acids enable the archaeal L7Ae box C/D RNP core protein to recognize and bind the K-loop RNA motif.RNA. 2010 Jan;16(1):79-90. doi: 10.1261/rna.1692310. Epub 2009 Nov 19. RNA. 2010. PMID: 19926724 Free PMC article.
-
Small non-coding RNAs in Archaea.Curr Opin Microbiol. 2005 Dec;8(6):685-94. doi: 10.1016/j.mib.2005.10.013. Epub 2005 Oct 26. Curr Opin Microbiol. 2005. PMID: 16256421 Review.
-
Small regulatory RNAs in Archaea.RNA Biol. 2014;11(5):484-93. doi: 10.4161/rna.28452. Epub 2014 Mar 31. RNA Biol. 2014. PMID: 24755959 Free PMC article. Review.
Cited by
-
Heat shock in C. elegans induces downstream of gene transcription and accumulation of double-stranded RNA.PLoS One. 2019 Apr 8;14(4):e0206715. doi: 10.1371/journal.pone.0206715. eCollection 2019. PLoS One. 2019. PMID: 30958820 Free PMC article.
-
Splicing Endonuclease Is an Important Player in rRNA and tRNA Maturation in Archaea.Front Microbiol. 2020 Nov 20;11:594838. doi: 10.3389/fmicb.2020.594838. eCollection 2020. Front Microbiol. 2020. PMID: 33329479 Free PMC article.
-
Selective Translation of Low Abundance and Upregulated Transcripts in Halobacterium salinarum.mSystems. 2020 Jul 28;5(4):e00329-20. doi: 10.1128/mSystems.00329-20. mSystems. 2020. PMID: 32723790 Free PMC article.
-
Identification of NAD-RNA species and ADPR-RNA decapping in Archaea.Nat Commun. 2023 Nov 21;14(1):7597. doi: 10.1038/s41467-023-43377-x. Nat Commun. 2023. PMID: 37989750 Free PMC article.
-
Directed evolution of orthogonal RNA-RBP pairs through library-vs-library in vitro selection.Nucleic Acids Res. 2022 Jan 25;50(2):601-616. doi: 10.1093/nar/gkab527. Nucleic Acids Res. 2022. PMID: 34219162 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

